Origins, natural reservoirs and Interspecies transmission of SARSCoV- 2 and other SARS-like CoVs (ONRITS)
 Project Coordinator:
Project Coordinator:
• Dr. Paul Brey, Director, Institut Pasteur du Laos, Vientiane, Lao PDR
• Dr. Khamsing Vongphayloth, Research entomologist, Institut Pasteur du Laos
Staff members:
• Vincent Lacoste, Head of Arbovirus & Emerging Viral Disease Laboratory, Institut Pasteur du Laos
• Somphavanh Somlor, Research Scientist, Arbovirus & Emerging Viral Disease Laboratory, Institut Pasteur du Laos
• Khaithong Lakeomany, Technician entomologist, Institut Pasteur du Laos
• Nothasine Phommavanh, Technician entomologist, Institut Pasteur du Laos
• Khammany Oudomsouk, Technician entomologist, Institut Pasteur du Laos
The participating team from the National University of Laos:
• Bounsavanh Daongboupha (Chiropteran, Faculty of Environmental Science)
• Vilakhan Xayaphet (Chiropteran Assistant, Faculty of Environmental Science)
• Daosavanh Sanamxay (Rodent, Faculty of Environmental Science)
Outside partners:
• National Institute of Hygiene & Epidemiology- Vietnam
• The Pathogen Discovery Laboratory, IP-Paris-France
Funded by the « URGENCE COVID-19 » fundraising campaign of Institut Pasteur, Institut Pasteur du Laos, the United Kingdom Foreign and Commonwealth Office as well as Luxembourg Development special grant.
Period of Project: 2020-2021
Summary
Since the discovery of severe acute respiratory syndromeassociated coronavirus (SARS-CoV) in Chinese horseshoe bats (Rhinolophus sinicus) in Hong Kong in 2005, a plethora of molecular epidemiological studies have been carried out in bats and animal species around the world to detect other animal reservoirs of this highly pathogenic CoV (Wong et al., 2019). Interestingly, of the >30 CoVs found in bats, a majority have been found in various species of horseshoe bats (Rhinolophus spp.) from China. A few others have been found in other bat genera such as, Aselliscus, Chaerephon, and Hipposideros.
Given the fact that two SARS Coronaviruses have emerged and caused important human loss of life in the past seventeen years (SARS and COVID-19), the scientific community postulates that other SARS Coronaviruses are presently circulating in Rhinolophus and other bat populations and could spill over into human populations like SARS and COVID-19; hence, the surveillance and identification of these novel putative coronaviruses is necessary to mitigate a new future pandemic.
A better scientific understanding of the origin, natural history and dispersal of SARS-CoV-2 and SARS-like- CoVs in their natural environment, as well as the mechanisms leading to their interactions and interspecies jumping: Bat-Bat; Bat-Animal and Bat-Human are paramount to mitigate future spillover events that result in devastating epidemics like the one we are witnessing today with SARS-CoV-2 in China and around the world.
Objectives
The specific objectives of this project are to:
• Assess the presence of SARS-CoV2 and related viruses in Laos and Vietnam wildlife (bats and other mammal communities at the interface with them);
• Search for other SARS-like CoVs using New Generation Sequencing (NGS)-based metagenomic analysis of selected samples;
• Detect past beta-coronavirus infections using specifically developed antibody tests for SARS-CoV2 and novel closely-related viruses.
Methodology
The field collection was conducted in Laos by teams from the Institut Pasteur du Laos (IP-Laos), and in Vietnam by the National Institute of Hygiene & Epidemiology (NIHE)-Vietnam, in collaboration with the Pathogen Discovery Laboratory at the Institut Pasteur (IP-Paris) to determine the presence of SARS-CoV-2 and to identify its natural host using Next Generation Sequencing (NGS), virus isolation, etc.
Site selection and time for field sampling in Laos
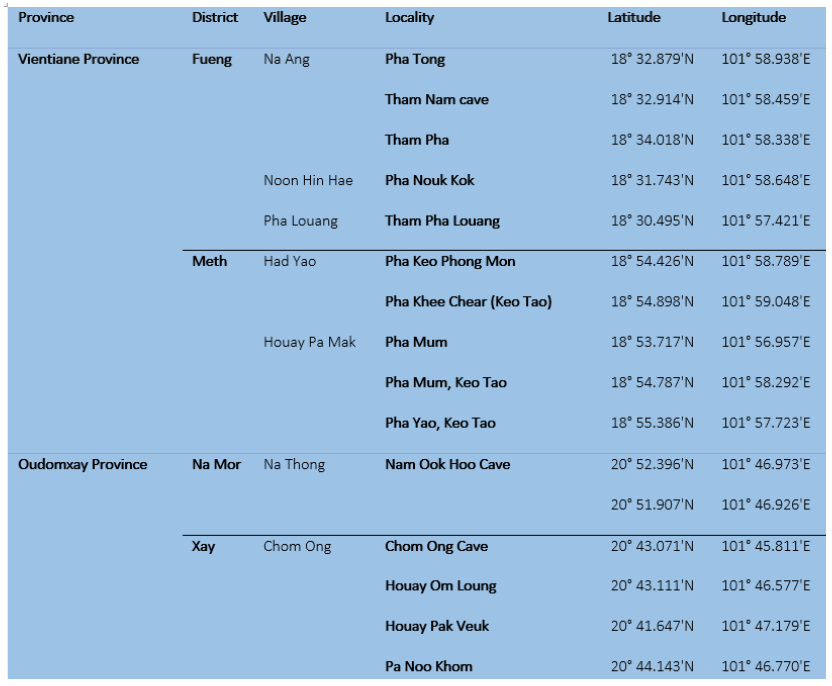
Between July 2020 and January 2021, our team conducted field collection from 4 sites within 2 provinces (Table 1): Fueng and Meth Districts, Vientiane Province, and in Namor and Xay Districts, Oudomxay Province. Our field mission was authorized and facilitated by the Ministry of Agriculture and Forestry of Laos.
Table 1: selection sites for our field collection between 2020-2021
Training on biosafety prior to field bat collection
The training prior to fieldwork was organized once for 6 participants who joined the fieldwork collection. The one-day training session was organized on 24 June 2020 at IPLaos. The aim was to introduce participants to our SOPs and to the transmission risks of infectious agents that may arise from bats/rodents and how to identify, assess, and mitigate the risks, as well as to make participants better understand methods that could minimize the risks, and to identify and practice the use of PPE suitable for the fieldwork.
Figure 1: Biosafety training at IP-Laos prior field investigation

Field collection and identification procedure
Bats
Bats were collected using four-bank harp traps (Francis, 1989) (Figure 2) and mist nets. Harp traps were set across natural trails and over small streams in the forest understorey, and at the entrance of caves in relatively concealed conditions. Mist nets were also set at similar places to harp traps but were more often used in open spaces. Both harp traps and mist nets were set before sunset; harp nets were left overnight. Captured bats were held individually in cloth bags. Species, sex, age (adult or juvenile), and reproductive condition (pregnant or lactating) were determined in the field. Species were identified following Francis (2008), Csorba et al. (2003), and Corbet and Hill (1992). Adults or juveniles were identified by the presence of unfused epiphyses of the phalanges and metacarpal joints (Brunet-Rossinni and Wilkinson, 2009). The reproductive status of female bats was determined by examining the nipples (Racey, 2009). Some external characteristics, including forearm length (FA), were measured using calipers and body mass (W) was taken using a Pesola spring balance. Most bats were marked with wing bands for individual identification and were released at the capture point within 12 hours. For each species, two or three specimens were retained to confirm identification. Bats were euthanized using chloroform. Specimens were fixed in 95% ethanol in the field and were transferred to 70% ethanol when they were brought back to the laboratory. All specimens were registered and cataloged in the Zoological Collection of the Faculty of Environmental Sciences, National University of Laos, Lao PDR.
Figure 2. Harp trap setting

Rodents
Rodent habitats were identified, especially at the entrance of caves and peri karstic areas. Small wire live-traps (14x14x30 cm in WxHxL), Sherman traps, or other similar devices specially fitted for small mammals were set for rodent collection. Between 50 and 100 traps per night were placed in the format of either grid or transect according to the topography, and each trapping session lasted from 6-8 nights. Traps were baited with appropriate baits (e.g., bananas, sticky rice, or dried fish) and checked every morning.
For each species of rodent, two or three specimens were retained to confirm identification. Rodents were euthanized using chloroform. Specimens were fixed in 95% ethanol in the field and were transferred to 70% ethanol when they are brought back to the laboratory. All of the specimens were registered and cataloged in the Zoological Collection of the Faculty of Environmental Sciences, National University of Laos, Lao PDR.
Types of biological samples from animals
The following basic set of samples was collected, if possible, from each bat and other animals:
• Oral-pharyngeal swab: 1 aliquot
• Feces (fresh fecal sample) or rectal swab: 1/2 aliquots
• Blood (serum; red blood cell/white blood cell pellet): 2 aliquots
• Urine (free catch method or urogenital swab): 1/2 aliquot
• Ectoparasites : 1 aliquot
Laboratory procedure
Extraction Total nucleic acids (DNA/RNA) were extracted from biosamples, especially for bat and rodent anal swabs by using Nuclo Spin®8 Virus (MACHEREY-NAGAL, Germany).
Lab techniques and assay procedure for Pan-corona screening at IP-Laos
cDNA was synthesized using Maxima H minus first-strand cDNA synthesis kit (Thermo Scientific) with random hexamers following the manufacturer’s instructions. Pan-corona PCR targeting the RNA-dependent RNA Polymerase gene was used for screening of coronaviruses as previously described by Chu et al. 2011. PCR products of the expected size were directly sequenced on both strands by Sanger sequencing using the nested PCR primers. The sequences obtained were confirmed by similarity analysis using the NCBI BLASTn search (http://www.ncbi.nlm.nih.gov/BLAST).
NGS at IP-Paris
Total nucleic acids (DNA/RNA) for both positive and negative samples and an aliquot of anal swabs that were positive following pan-corona PCR at IP-Laos were sent to IP-Paris for in-depth analysis.
Preliminary results
Biological sampling A total of 645 bats were captured from 4 collection sites and 608 saliva, 539 anal/feces, 157 urine swabs and 246 blood samples were collected. In addition, ectoparasites were collected from 74 bats (Table 2).
Table 2: Total number of bats captured and biological samples
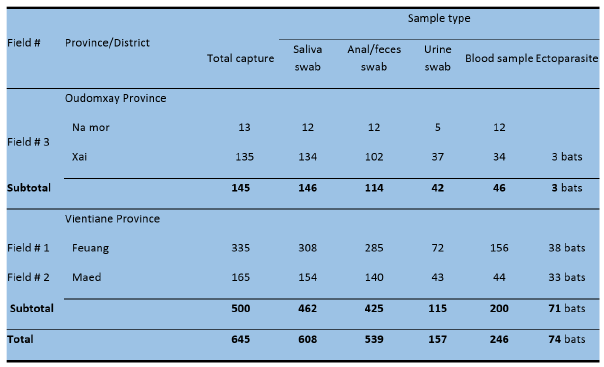
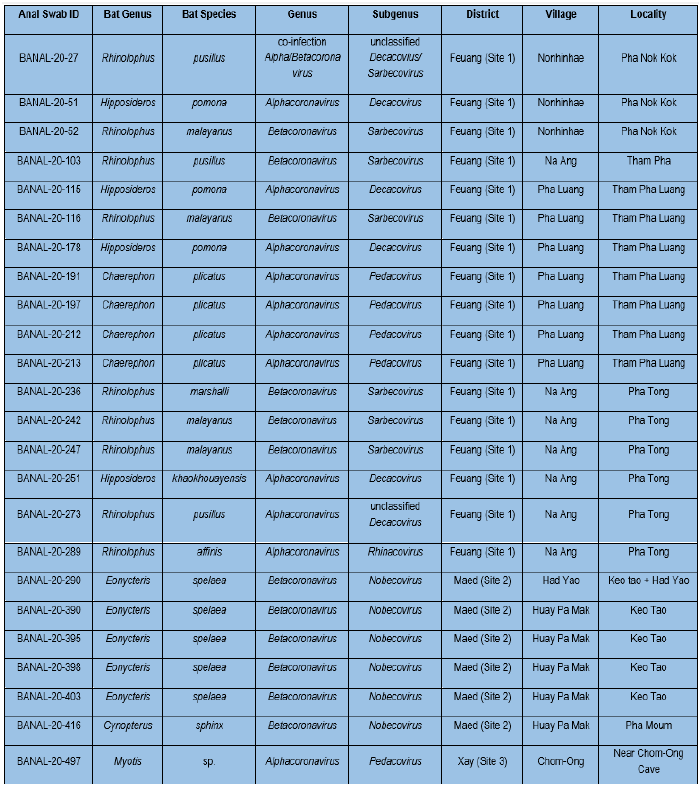
Table 4: Results from pan-coronavirus RT nested PCR and Sanger sequencing for bat anal swabs/feces at IP-Laos. Blast search based on nucleotide sequences.
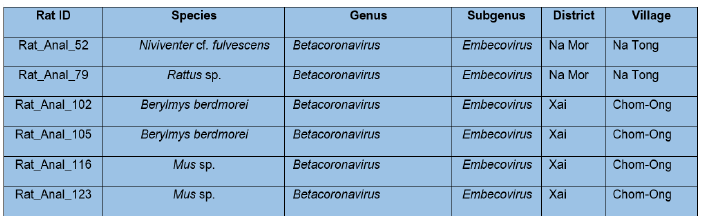
For rodents, a total of 115 extracted products of bat anal swabs/faces were screened at IP-Laos by targeting the RNA-dependent RNA polymerase gene using a pan-coronavirus RT nested PCR approach. A total of 6 individuals from 4 rodent species were positive (Table 5). BLAST analysis of obtained sequences from Sanger sequencing identified Betacoronavirus sequences of the Embecovirus subgenus. Six sequences of the Embecovirus subgenus were identified from 2 Berylmys berdmorei, 2 Mus spp., 1 Niviventer cf. fulvescens, and 1 Rattus sp. (Table 5).
Table 5: Results from pan-coronavirus RT nested PCR and Sanger sequencing for rodent anal swabs/feces at IP-Laos. Blast search based on nucleotide sequences.
NGS preliminary results from bat anal swabs/feces samples at IP-Paris
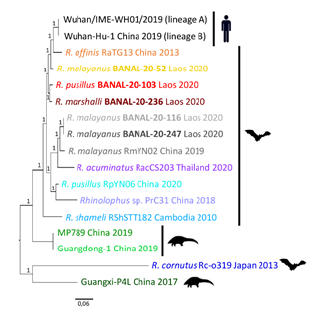
The subgenus of Sarbecoviruses was selected as priority for complete genome sequencing as SARS-CoV-2 belongs to this genus. A total of 5 complete genome sequences were obtained by NGS from seven Sarbecovirus positives by pan-coronavirus PCR. BLAST analysis of obtained complete sequences showed that these 5 sequences from three bat species – Rhinolophus malayanus, R. marshalli, and R. pusillus had identity >96% to SARS-CoV-2 (Table 6). Phylogenetic analyses of the complete genome sequences of human SARS-CoV-2, and other bat and pangolin sarbecoviruses showed that the 3 Laotian bat coronaviruses: R. malayanus BANAL-20-52, R. pusillus BANAL-20-103, and R. marshalli BANAL-20-236 were closely related to R. affinis RaTG13 coronaviruses and human SARS-CoV-2, while 2 other: R. malayanus BANAL-20-116 and BANAL-20-247 coronaviruses were grouped to a sister clade with other bat coronaviruses (RmYN02, RacCS203, RpYN06, and PrC31) from different Rhinolophus species (Figure 3). Like other sarbecoviruses found so far in animals from the Asian Peninsular region, all Laotian bat sarbecoviruses found here had an absence of the furin cleavage site.
Table 6: Results from complete genome sequencing at IP-Paris. Blast search based on nucleotide sequences.
Figure 3. Phylogenetic analyses of the complete genome sequences of Laotian bat coronaviruses, human SARSCoV- 2, and other bat and pangolin sarbecoviruses
Other preliminary results were obtained from IP-Paris teams investigating molecular dynamics simulations of the binding of Laotian bat-sarbecovirus RBD to human ACE2 complexes and Pseudoviruses expressing BANAL-20-236 spike entry into cells express human ACE2. Laotian bat-sarbecovirus RBD was able to bind to human ACE2 and Pseudoviruses expressing BANAL-20-236 spike could enter into cells expressing human ACE2.
Conclusion/Ongoing activities/Perspective
Our study showed that 25 coronaviruses were detected from 10 bat species (out of 539 samples tested), of which 3/5 sarbecoviruses with full-length sequences were closely related to SARS-CoV-2 > 96%. Preliminary results showed that Laotian bat-sarbecovirus RBD was able to bind to human ACE2 through modeling and binding experiments, and Pseudoviruses expressing BANAL-20-236 spike could enter into cells expressing human ACE2.
Further experiments such as investigating whether Pseudoviruses expressing BANAL-20-236 spike can directly enter into human cells, and pathogenicity of these viruses are warranted. Other questions will need also to be addressed such as whether the bat-exposed local populations have been infected by one of these viruses in Laos, whether such infections were associated with any symptoms, and whether exposure can confer immunity against subsequent SARS-CoV-2 infection.
Our preliminary results showed that there exist other bat sarbecoviruses that seem to have the same potential for infecting humans in Laos. People with close contact with bats seem to be at risk of being exposed, so they will need some level of personal protection if they have contact with the bats.
References
Brunet-Rossinni, A.K. and G.S. Wilkinson. 2009. Methods for age estimation and the study of senescence in bats. Pp: 315-325, in Ecological and behavioral methods for the study of bats, 2nd edition (T.H. Kunz and S. Parsons, eds). The Johns Hopkins University Press, Baltimore, xvii + 901 pp.
Chu, D. K. W. et al. Avian Coronavirus in Wild Aquatic Birds. Journal of Virology 85, 12815–12820 (2011).
Chua B.K., et al. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes and Infection, 4: 145–151.
Corbet, G.B. and J.E. Hill. 1992. The mammals of the Indomalayan region: a systematic review. Natural History Museum Publications and Oxford University Press, 488 pp.
Csorba, G., P. Ujhelyi, and N. Thomas. 2003. Horseshoe bats of the World (Chiroptera: Rhinolophidae). Alana Books, xxxii + 160 pp.
Francis, C.M. 1989. A comparison of mist nets and two designs of harp traps for capturing bats. Journal of Mammalogy, 70: 865-870.
References 37 INSTITUT PASTEUR DU LAOS ANNUAL REPORT 2021-2022
Francis, C.M. 2008. A field guide to the mammals of Thailand and South-east Asia. New Holland Publishers (UK) Ltd and Asia Books Co., Ltd, Bangkok, Thailand, 392 pp.
Racey, P.A. 2009. Reproductive assessment of bats. Pp: 249-264, in Ecological and behavioural methods for the study of bats, 2nd edition (T.H. Kunz and S. Parsons, eds). The Johns Hopkins University Press, Baltimore, xvii + 901 pp.
Smith,C., C. DeJong and H.E. Field (2010) Sampling small quantities of blood from microbats. Acta Chiropterologica. 12(1): 255-258.Bates, P.J.J. and D.L. Harrison. 1997. Bats of the Indian Subcontinent. Harrison Zoological Museum, Sevenoaks, Kent, England, xvi + 258 pp.
Wong ACP, Li X, Lau SKP, Woo PCY. Global Epidemiology of Bat Coronaviruses. Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. PMID: 30791586; PMCID: PMC6409556.