Arboviruses surveillance in Laos
 Collaborations.
Collaborations.
Twenty two hospitals from 11 provinces in Laos.
National Centre for Laboratory and Epidemiology (NCLE), Ministry of Health, Lao PDR.
Department of Communicable Diseases Control (DCDC), Ministry of Health, Lao PDR.
Funding.
Agence Française du Développement (AFD, Ecomore2 project), France.
Defense Threat Reduction Agency (DTRA), USA (Arboshield Plus project).
French Ministry of Higher Education, Research and Innovation (MESRI), France.
Objective.
The main objective of the arboviruses surveillance implemented at IPL since 2012 is to provide to the Lao Ministry of Health (MOH) information on circulation within Laos of the various arboviruses (particularly dengue and chikungunya viruses).
Background.
Since 2012, the Virology laboratory has been providing the Lao MOH with information on the circulation of arboviruses within Laos. To achieve this goal, the Virology laboratory has established a surveillance network for arboviruses in the Vientiane capital. In 2015, at the request of the MOH, surveillance was extended to the 2 southern provinces of Laos (Saravane and Attapeu). Since 2018, it has been extended to an additional 7 provinces (Champasack, Savannakhet, Luang-Prabang, Xiengkhuang, Phongsaly, Oudomxay and Vientiane province) through the implementation of Arboshield project, a collaboration with the Lao Army Health Service. As a result, the arbovirus surveillance network of IPL now includes a total of 22 hospitals (Figure 1).
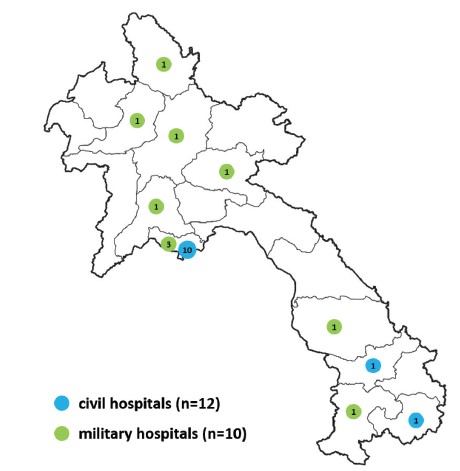
Figure 1: Map of IPL hospitals network contributing to the arboviruses surveillance. Civil hospitals (n=12) are indicated in blue and military hospitals (n=10) in green.
Methodology.
Samples.
Patient’s blood samples are collected on EDTA tubes and stored at +4°C prior to transportation to IPL for diagnosis and molecular characterization. At IPL, blood specimens are centrifuged and plasma samples are used to perform the various virological tests.
Virus, antigen and antibody detection
Virus nucleic acids are extracted from human plasma by NucleOspin RNA virus kit (Macherey-Nagel); NucleOspin 8 virus kit (Macherey-Nagel) or Nucleic Acid Extraction Rapid kit (Bioperfectus) according to the manufacturer’s instructions.
Extracted RNAs are then screened for the presence of dengue (DENV) and Zika (ZIKV) viruses by a duplex one-step real-time RT-PCR assay (Ou et al. 2020). The DENV serotype identification is performed by using a multiplex one-step real-time RT-PCR assay (Ou et al. 2020).
The plasma samples that test negative for DENV by PCR are screened by the Dengue NS1 Antigen + Antibodies Combo rapid test (SD Bioline) for detection of DENV NS1, anti-DENV IgM and anti-DENV IgG.
Finally, the samples that test negative for DENV by PCR and/or DENV NS1 by rapid test are then screened for the detection of chikungunya virus (CHIKV) using a realtime RT-PCR assay (Pastorino et al. 2005).
Genotyping
The sequencing of the envelope (E) gene (1485 nucleotides) is performed on representative DENV strains. The selection is based on the geographical origin of the patient, the year of collection, the Ct value measured by the pan-dengue real-time RT-PCR using primer sets previously described (Calvez et al. 2020, 1; Calvez, Bounmany, et al. 2021). The PCR products are purified with ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific) and sequenced using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on a 3500xL Genetic Analyzer apparatus (Applied Biosystems).
The raw sequences are analyzed and edited using Unipro UGENE software (Okonechnikov et al. 2012). Multiple sequence alignments are constructed with other sequences of publicly available representative DENV-1 or DENV-2 strains downloaded from GenBank (www. ncbi.nlm. nih.gov/nucleotide/), using the ClustalW program. Each alignment is checked manually. For phylogenetic analysis, maximum likelihood (ML) trees were constructed using MEGA version 10 (www. megasoftware.net), with a General Time Reversible (GTR) model with Gamma Distributed with Invariants Sites (G+I) and then edited with FigTree v1.4.4 software (http://tree.bio.ed.ac.uk/).
Results.
Dengue surveillance.
Between the 1st of January and 15th of October 2023, 1966 suspected cases were investigated at IPL. The majority of the samples received originated from Vientiane Capital hospitals (72%, n=1416). In total, 57% of the samples were provided by civil hospitals (n=1121) and 43% by military hospitals (n=845) (Figure 2).
Among the 1966 samples analysed at IPL, 62.5% were “confirmed acute dengue infection cases” (positive by RT-PCR or for dengue NS1), 2.4% were considered as “acute or recent dengue infection” (presence of anti-dengue IgM), 4.4% were considered as “inconclusive” (only presence of anti-dengue IgG) and in 28.4% of them we found no evidence of dengue virus infection (Figure 3).
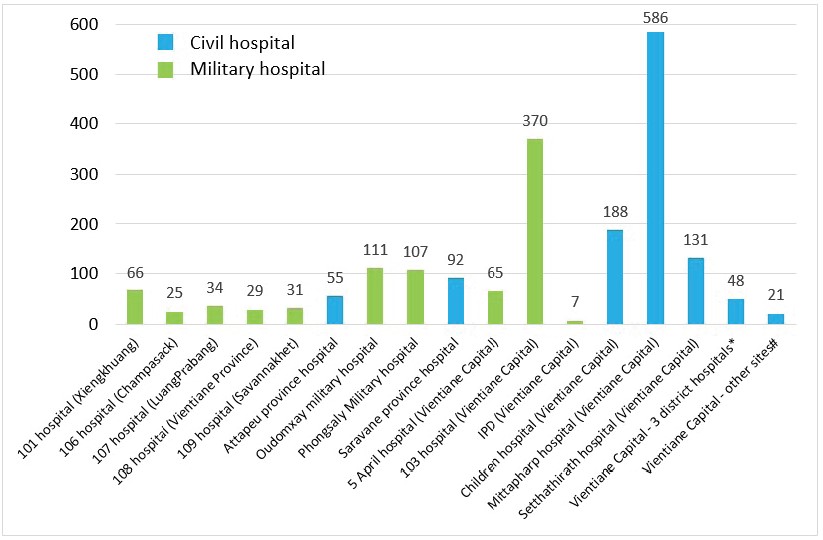
Figure 2: Origin of the samples collected for the arbovirus surveillance, received at IPL between 01st January and 15th October 2023. IDP: Institute of Disease Prevention of Lao Army; #: 3 district hospitals in Vientiane capital: Sisattanak district hospital (n=29); Hadxayfong district hospital (n=9); Xaythany district hospital (n=10). *: Other sites in Vientiane capital: French Medical Centre (n=13); IPL (n=7); Lao-NCLE/Bokeo provincial hospital (n=1).
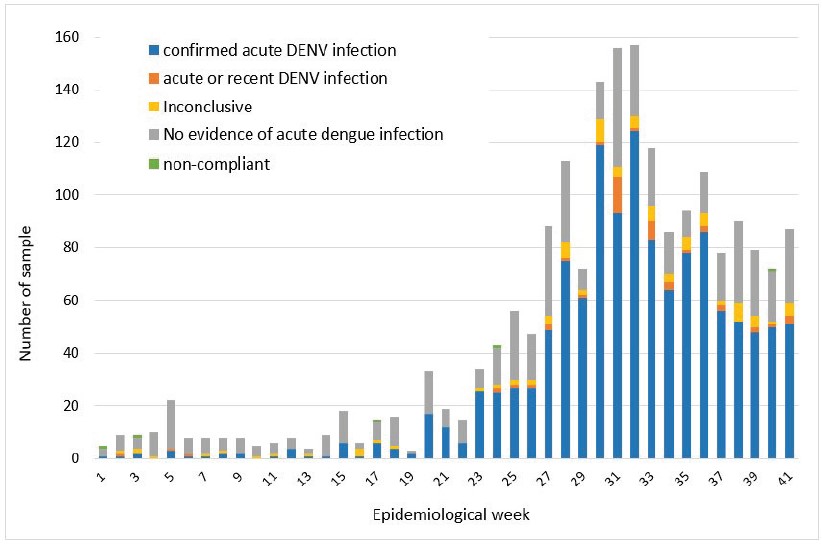
Figure 3: Conclusions of dengue diagnosis made on samples tested at IPL on a weekly basis between 01st January 2023 and 15th October 2023.
Serotype identification.
Between 1st January and 15th October 2023, dengue serotyping was performed on all 1249 samples positive by DENV PCR and serotyping results were obtained for more than 98,5% of them (n=1233). At the national level, DENV-2 was the predominant circulating serotype in the country representing 55.5% of all PCR-positive cases and the DENV-1 serotype was detected in 43.9% of cases. 7 cases of co-infection by DENV-1 and DENV-2 and 1 case of DENV-4 serotype were also detected in 2023.
Differential diagnosis.
The algorithm used to investigate dengue suspected cases at IPL includes a differential diagnostic approach. Since 2023, all samples received at IPL were screened by a duplex RT-PCR to detect DENV and ZIKV. The algorithm used to investigate dengue suspected cases at IPL includes a differential diagnostic approach. Since 2023, all samples received at IPL were screened by a duplex RT-PCR to detect DENV and ZIKV. Moreover, all samples that tested negative by DENV/ZIKV PCR were then tested for CHIKV infection by RT-PCR.
Genotyping of DENV-1 and DENV-2 strains detected during the 2020-2022 period.
In 2023, the laboratory characterized the genotype of 51 DENV samples identified between 2020 and 2022 (Figure 4): (i) 27 DENV-1 samples from Vientiane Capital and Attapeu, Champasack, Luang Prabang, Oudomxay, Phongsaly, Saravane, Vientiane, Xaysomboune and Xiengkhuang provinces and (ii) 24 DENV-2 samples from Vientiane Capital and Attapeu, Champasack, Luang Prabang, Oudomxay, Saravane, Savannakhet and Vientiane provinces.
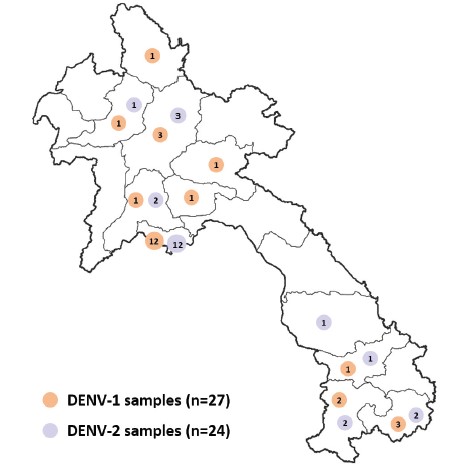
Figure 4: Origin of DENV-1 and DENV-2 samples selected for genotype analysis. DENV-1 samples (n=27) are indicated in orange and DENV-2 samples (n=24) in purple.
Detailed results can be found in the paper version of this report and in upcoming publications.
Discussion.
In 2023, Laos faced an important outbreak of dengue fever with 30,751 dengue suspected cases reported until week 41, including 18 fatal cases (Dengue NCLE report – 15 October 2023). During the same period, the blood specimens of 1966 patients with suspected dengue were tested at IPL. Among them, 1268 (62.5%) cases were classified as “confirmed acute dengue infection”, 48 (2.4%) as “acute or recent dengue infection”, 87 (4.4%) as “inconclusive” and 558 (28.4%) cases, there was no evidence of dengue infection. In 2022, Laos also faced an important outbreak of dengue fever with 39,091 dengue suspected cases and 24 fatal cases. In contrast, during 2020 and 2021, significantly fewer dengue suspected cases were reported nationally (8217 and 1683 suspected cases, respectively).
The proportion of confirmed acute dengue infection cases (samples positive by DENV RT-PCR or for dengue NS1) found among the samples obtained in 2023 (62,5%) is comparable with the proportion of confirmed cases observed during the previous years (50% to 61% between 2020 and 2022).
In 2023, a change in the serotype circulation was observed in the country. Indeed, DENV-2 became the predominant serotype circulating, representing 55.5% of all PCR-positive cases and the DENV-1 serotype was detected in 43.9% of cases.
In 2023, we determined the genotype of 51 DENV strains detected between 2020 and 2022 (27 DENV-1 and 24 DENV-2) originating from 11 Lao provinces, by sequencing the complete E gene.
Zika virus (ZIKV) was also identified as a possible threat in Laos and was already detected in neighbouring countries such as Cambodia, Thailand and Vietnam (Lim, Lim, et Yoon 2017). In February 2020, a baby was born with microcephaly in Vientiane. The medical, epidemiological and laboratory investigations demonstrated that this was the first probable congenital Zika case reported in Laos (Calvez, Vetsaphong, et al. 2021). This is why, the algorithm used to investigate dengue suspected cases at IPL includes CHIV and ZKV in the differential diagnosis. Since 2023, all samples received at IPL were screened by a duplex RT-PCR to simultaneously detect DENV and ZIKV (Ou et al. 2020). Moreover, all samples that test negative for DENV by PCR are then tested for CHIKV infection by RT-PCR (Pastorino et al. 2005).
Conclusion and perspectives.
In 2023, 1268 samples were classified as “confirmed acute dengue infection” (62.5%), 48 as “acute or recent dengue infection” (2.4%) among the 1966 samples obtained from patients presenting with dengue-like symptoms at the hospitals participating in the surveillance system.
All results of the arboviruses surveillance were shared i) on a daily basis with clinicians; ii) on a weekly basis with NCLE and DCDC; and iii) on a monthly basis with DCDC. NCLE and DCDC consolidate testing data from all laboratories and report aggregated data to the MOH. This active surveillance enables the MOH to track the epidemiologic situation of dengue in Laos.
The implementation of a new sequencing method (Next Generation Sequencing) will allow more in-depth genomic characterization of DENV and CHIKV in order to better understand the dynamics of circulation of these viruses in Laos.
References.
Balière, Charlotte, Elodie Calvez, Jean-Michel Thiberge, Somphavanh Somlor, Mathias Vandenbogaert, Marc Grandadam, et Valérie Caro. 2023. « A Six Years (2010- 2016) Longitudinal Survey of the Four Serotypes of Dengue Viruses in Lao PDR ». Microorganisms 11 (2):243. https://doi.org/10.3390/microorganisms11020243.
Calvez, Elodie, Phaithong Bounmany, Charlotte Balière, Somphavanh Somlor, Souksakhone Viengphouthong, Thonglakhone Xaybounsou, Sitsana Keosenhom, et al. 2021. « Using Background Sequencing Data to Anticipate DENV-1 Circulation in the Lao PDR ». Microorganisms 9 (11): 2263. https://doi.org/10.3390/ microorganisms9112263.
Calvez, Elodie, Phaithong Bounmany, Somphavanh Somlor, Thonglakhone Xaybounsou, Souksakhone Viengphouthong, Sitsana Keosenhom, Paul T. Brey, Vincent Lacoste, et Marc Grandadam. 2022. « Multiple Chikungunya Virus Introductions in Lao PDR from 2014 to 2020 ». PloS One 17 (7): e0271439. https://doi. org/10.1371/journal.pone.0271439.
Calvez, Elodie, Somphavanh Somlor, Souksakhone Viengphouthong, Charlotte Balière, Phaithong Bounmany, Sitsana Keosenhom, Valérie Caro, et Marc Grandadam. 2020. « Rapid Genotyping Protocol to Improve Dengue Virus Serotype 2 Survey in Lao PDR ». PloS One 15 (8): e0237384. https://doi.org/10.1371/ journal.pone.0237384.
Calvez, Elodie, Phommady Vetsaphong, Somphavanh Somlor, Thonglakhone Xaybounsou, Souksakhone Viengphouthong, Myrielle Dupont-Rouzeyrol, Virginie Pommelet, et Paul T. Brey. 2021. « First Probable Case of Congenital Zika Syndrome in Lao People’s Democratic Republic ». International Journal of Infectious Diseases: IJID: Official Publication of the International Society for Infectious Diseases 105 (avril): 595‑97.
Lim, Sl-Ki, Jacqueline Kyungah Lim, et In-Kyu Yoon. 2017. « An Update on Zika Virus in Asia ». Infection & Chemotherapy 49 (2): 91‑100. https://doi.org/10.3947/ ic.2017.49.2.91.
Okonechnikov, Konstantin, Olga Golosova, Mikhail Fursov, et UGENE team. 2012. « Unipro UGENE: A Unified Bioinformatics Toolkit ». Bioinformatics (Oxford, England) 28 (8): 1166‑67. https://doi.org/10.1093/ bioinformatics/bts091.
Ou, Tey Putita, Chanvannak Yun, Heidi Auerswald, Saraden In, Rithea Leang, Rekol Huy, Rithy Choeung, Philippe Dussart,Veasna Duong. 2020. « Improved Detection of Dengue and Zika Viruses Using Multiplex RT-qPCR Assays ». Journal of Virological Methods 282 (août): 113862. https://doi.org/10.1016/j. jviromet.2020.113862.
Pastorino, Boris, Maël Bessaud, Marc Grandadam, Severine Murri, Hugues J. Tolou, et Christophe N. Peyrefitte. 2005. « Development of a TaqMan® RT-PCR Assay without RNA Extraction Step for the Detection and Quantification of African Chikungunya Viruses ». Journal of Virological Methods 124 (1‑2): 65‑71. https:// doi.org/10.1016/j.jviromet.2004.11.002.
Somlor, Somphavanh, Khamsing Vongpayloth, Laure Diancourt, Philippe Buchy, Veasna Duong, Darouny Phonekeo, Pakapak Ketmayoon, et al. 2017. « Chikungunya Virus Emergence in the Lao PDR, 2012-2013 ». PloS One 12 (12): e0189879. https://doi. org/10.1371/journal.pone.0189879.






