Ectoparasites and pathogen surveillance in Laos

Collaborations
U.S. Naval Medical Research Unit-2
Lao-Oxford University-Mahosot Hospital-Wellcome Trust Research Unit (LOMWRU)
Pathogen Discovery Laboratory, Institut Pasteur, Paris, France
Funding
U.S. Naval Medical Research Center-Asia (NMRC-A) in support of the Department of Defense Global Emerging Infections Surveillance and Response System (DoDGEIS)
Objectives
The objectives of the project are 1) to collect from study areas up to 10,000 ticks and other associated arthropod specimens, 2) to perform morphological identification (ID) of indigenous ticks and other associated arthropods by combining morphological and molecular identification of vector DNA for submission and development of a regional repository, 3) to build local capacities and competencies.
Background
Vector-borne diseases constitute a significant infectious disease risk for deployed military personnel and for local populations. In Laos, definitive diagnosis is often not available for vector-borne illnesses, so the infectious diseases that are a threat to military and civilian populations are not well-defined. In order to identify common and emerging vector-borne pathogens in Laos, the Institut Pasteur du Laos (IP-Laos) in collaboration with NAMRU-2 Singapore (SG) and other partners has established a study to assess the distribution and the infection potential of vectors (including ticks and associated arthropods). In this study, ticks and associated arthropod vectors were collected from the environment and their associated hosts. They were transported to IP-Laos’ laboratory in Vientiane, where a wide range of diagnostic tests were performed to identify both the vectors and their associated pathogens.
Methodology
Working group
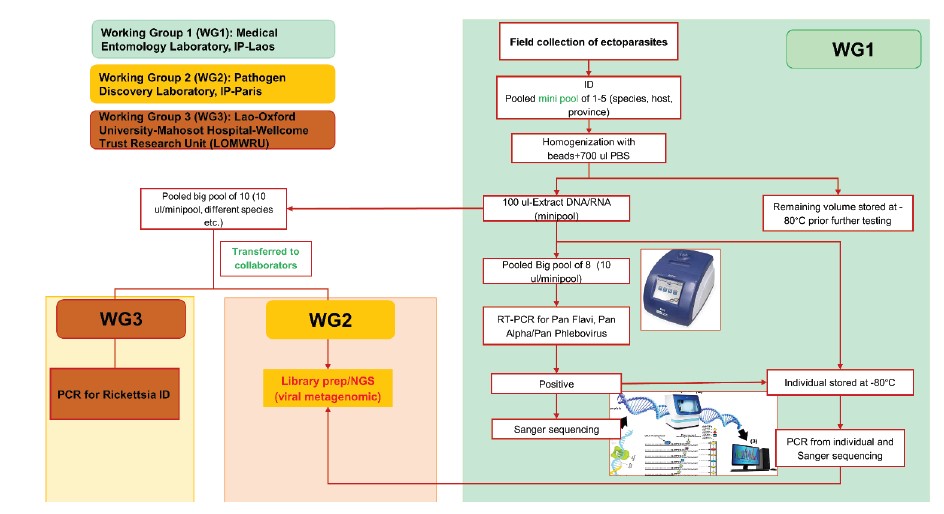
In order to identify common and emerging vector-borne diseases caused by viruses and rickettsia in Laos, this study was conducted between 2022–2023 in collaboration with two main teams with high levels of expertise in this field: the Pathogen Discovery Laboratory at Institut Pasteur Paris, and the Lao-Oxford University-Mahosot Hospital- Wellcome Trust Research Unit (LOMWRU) (Fig. 1).
Field sites
We completed five field collection missions: two in Phongsaly and three in Borlikhamxay (fig. 2).
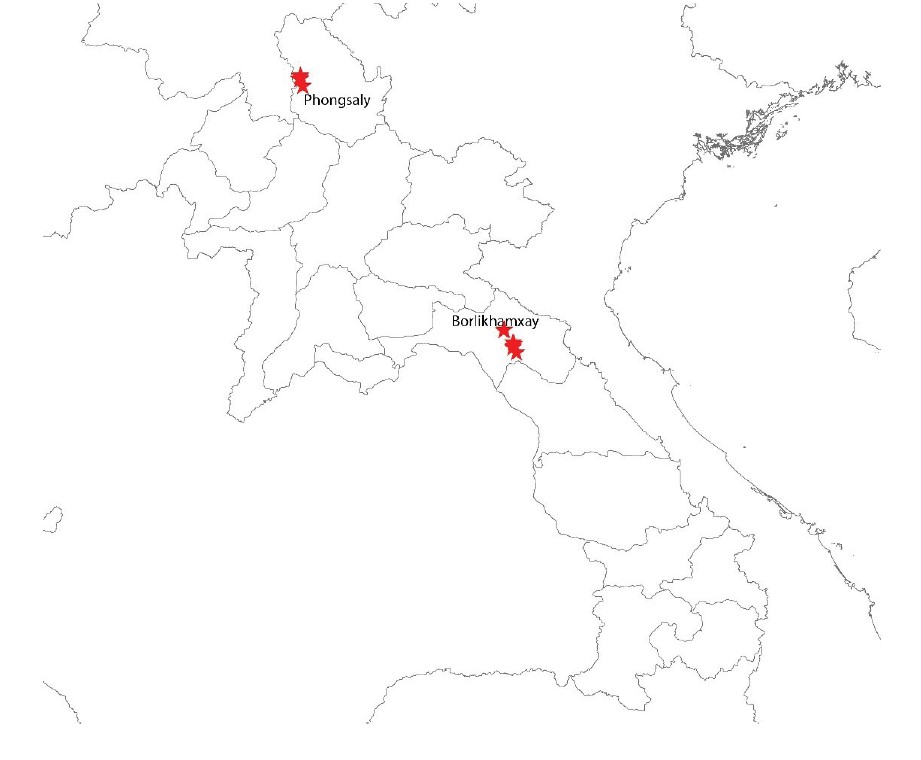
Figure 2: Sampling sites in Phongsaly and Borlikhamxay provinces.
Field collection procedures
Tick dragging/flagging: tick dragnets were swept/dragged along the forest ground at approximately 1–2 m intervals before being examined for the presence of ticks (Fig. 3). Ticks were removed from the sheets using forceps, then transferred to 1.5 ml labeled cryotubes, and stored -20°C. Our dragnet collection was carried out on all three sites.
Figure 3: Tick dragging in the forest in Phongsaly province.


Small mammals trapping: in each study site, 50 cage traps (baited with bananas, sticky rice, or dried fish/meat) were placed in a plantation or forest in the format of a transect according to the topography (Fig. 4).
Figure 4: Setting cage traps for rodent capture in forest areas.

Additional tick collection was carried out by examining domestic animals (cows). The animal owners were asked to help to examine their animals. Once ticks attached on animals were found, they were collected by direct hand removal with forceps.
All samples were stored at -20°C in the field and transported to IPL using dry ice.
Laboratory work
Identification of ectoparasites: ticks were identified and grouped under microscopes in cooling conditions (on ice packs) by using reference determination from Dr. Richard G. Robbins of the US Armed Forces Pest Management Board (AFPMB), together with related references from Southeast Asia, Japan, Korea, the Ryukyu Islands by Yamaguti, Tipton et al.1, L. E. Robinson keys for genus Amblyomma by Nuttall, Cooper et al.2, and keys from Thailand by Tanskull and Inlao3 for adult Haemaphysalis ticks. As there are no morphological identification keys available for pre-imago forms, all larval and nymph stages were grouped into genus. After tick identification and pooling, all information was registered with the Pathogen Asset Control System (PACS) software and all tick samples were stored at −80°C in Institut Pasteur du Laos for further analysis.
Chigger mites from rodents were mounted on the slide using a PVA mounting medium. Mite samples were identified using the compound microscope to genus level by referring to the published taxonomic key of Nadchatram & Dohany 19744.
Sample preparation and RNA/DNA extraction: samples were pooled into “minipool” from 1-10 ticks/ectoparasites according to collection source, species, development stage, blood meal, and site. All the pools were extracted as following procedure: specimens were placed in a 1.5 ml vial containing 1 ml of 1X cold Phosphate Buffered Saline (PBS) and Lysing Matrix A zirconium beads (MP Biomedicals). Tick pools were homogenized for 10 min at a vibration frequency of 25/s in a TissueLyser II system (Qiagen). After grinding, beads and tissues were spun down by centrifugation for 5 min at 3000 rpm. To obtain total nucleic acid (both DNA and RNA) for bacterial and viral detection by polymerase chain reaction (PCR), 100 μl of each pool was extracted and purified by using the NucleoSpin® 8 Virus extraction kit following the manufacturer’s protocol. The remaining 400 μl of each pool was stored at –80°C for future pathogen isolation.
Arbovirus screening: samples were screened for the presence of phleboviruses and flaviviruses by means of conventional nested RT-PCR as previously described by Sanchez-Seco, Rosario et al. 20035; and Sanchez-Seco, Rosario et al. 20056.
Bacteria screening: the screening for bacteria was carried out by LOMWRU, based at Mahosot Hospital, Vientiane. To investigate the occurrence of Rickettsia spp. in ticks, a molecular screening approach targeting the 17kDa gene was taken (Jiang et al.7). For Rickettsia screening, a total of 1,272 ectoparasites from 678 small pools were retooled into 177 big pools according to collection sources, species, location, etc.
Next generation Sequencing (Institut Pasteur, Paris): a total of 1,063 ectoparasites collected from two provinces and from different collection methods were pooled into 36 big pools from 654 mini pools according to collection sources, species, location, etc., and sent to IP in Paris for NGS analysis.
In brief, at the Pathogen Discovery lab (IP-Paris), the quality and concentration of large pools were evaluated using Agilent 2100 Bioanalyzer and Qubit 2.0 Fluorometer (Invitrogen, USA) respectively. Total RNA libraries were constructed using the SMARTer Stranded Total RNA-seq Kit v3-Pico input mammalian (TaKaRa). DNA library quality and concentration were evaluated using the same techniques mentioned above. RNA sequencing was performed on the Illumina NextSeq2000 sequencer, in paired-end 2 x101 pb format (to achieve approximately 50 million reads), at the Biomics platform (Institut Pasteur, Paris). Bioinformatics analyses were carried out using a bioinformatics pipeline (Microseek) developed by LDP team (see https://pubmed.ncbi.nlm. nih.gov/36146797/).
Results
A total of 3,621 ticks and other ectoparasites were collected from five field missions in two provinces, of which 1,610 were from Phongsaly and 2,011 were from Borlikhamxay. Ticks were classified into 15 species and 5 genera.
A total of 1,203 pools containing 2,372 ticks and 1 louse were extracted and screened by Pan-flaviviruses and Pan-phleboviruses PCRs at IPL.
A total of 1,063 ectoparasites collected from two provinces and from different collection methods were pooled according to collection sources, species, location, etc., in 36 big pools and sent to Institut Pasteur in France for NGS analysis. The analysis of the sequences generated is now ongoing.
A total of 30/177 big pools (265/1,272 samples) tested positive for Rickettsia spp. by PCR on the 17KDa gene.
More detailed results will be provided in an upcoming publication.
Discussion
A total of 3,621 ticks and other ectoparasites were collected from five field missions in two provinces. Ticks were classified into 15 species of 5 genera. As there are no morphological keys for identifying ticks at larval and nymphal stages, the association between adult ticks and their imago stages using molecular techniques is needed. Results from specific RT-PCR on Pan-flaviviruses and Pan-phleboviruses were all negative. However, these primer sets were designed only for some known virus identification; the analysis of metagenomic results generated by NGS should provide more information on viral pathogens associated with ticks in Laos. Our results from Rickettsia screening also highlight the importance of ticks and chigger mites as vectors of rickettsia diseases in Laos.
Conclusion & perspectives
Laotian tick fauna is more diverse than anticipated, and much remains to be discovered. LOMWRU will continue the work on Rickettsia species identification. The Institut Pasteur in Paris will continue to work on metagenomic analysis.
References
1. Yamaguti N., V. J. Tipton, H. L. Keegan, and S. Toshioka (1971). Ticks of Japan, Korea, and the Ryukyu islands. Brigham Young University Science Bulletin, Biological Series 15(1):1.
2. Nuttall, G. H. F., W. F. Cooper, C. Warburton, L. E. Robinson, and D. R. Arthur. 1926. Ticks: pt. IV. The genus Amblyomma. Cambridge University Press.
3. Tanskull, P. and I. Inlao. 1989. Keys to the adult ticks of Haemaphysalis Koch, 1844, in Thailand with notes on changes in taxonomy (Acari: Ixodoidea: Ixodidae). J Med Entomol 26(6): 573–600.
4. Nadchatram, M. and A. L. Dohany. 1974. A pictorial key to the subfamilies, genera and subgenera of Southeast Asian chiggers (Acari, Prostigmata, Trombiculidae). Bulletin from the Institute for Medical Research Federation of Malaysia, 16: 1–67.
5. Sanchez-Seco, M.P., et al., Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J Med Virol, 2003. 71(1): p. 140-9.
6. Sánchez-Seco MP, Rosario D, Domingo C, Hernández L, Valdés K, Guzmán MG, Tenorio A. Generic RT-nested- PCR for detection of flaviviruses using degenerated primers and internal control followed by sequencing for specific identification. J Virol Methods. 2005 Jun;126(1- 2):101-9. doi: 10.1016/j.jviromet.2005.01.025. PMID: 15847925.
7. Jiang J., Chan T., Temenek J.J., Dasch G.A., Ching W. and Richards A.L. (2004). Development of a Quantitative Real time Polymerase Chain Reaction Assay Specific for Orientia tsutsugamushi. American Journal of Tropical Medicine and Hygiene, 70(4): 351-356







