Investigation and Response to Emerging Viral Pathogens (IREVP): Pilot phase

Project Coordinator:
• Paul Brey, Director, Institut Pasteur du Laos, Vientiane, Lao PDR
• Somphavanh Somlor, Research Scientist, Virology and pathogen discovery laboratory
• Chittaphone Vanhnollat, Research Scientist, Virology and pathogen discovery laboratory
• Khamsing Vongphayloth, Research entomologist
IP-Laos team:
• Khaithong Lakeomany, Technician entomologist
• Nothasine Phommavanh, Technician entomologist
• Khanmany Oudomsouck, Technician entomologist
• Tavun Pongsanarm, Trainee on zoology
• Phaithong Bounmany, Technician, Virology and pathogen discovery laboratory
• Souksakhone Viengphouthong, Technician, Virology and pathogen discovery laboratory
Local partners: National University of Laos:
• Bounsavanh Daongboupha (Chiropteran, Faculty of Environmental Science)
• Vilakhan Xayaphet (Chiropteran Assistant, Faculty of Environmental Science)
• Daosavanh Sanamxay (Rodent, Faculty of Environmental Science)
National Animal Health Laboratory:
• Phouvong Phommachanh, National Animal Health Laboratory, Ministry of Agriculture and Forestry
• Watthana Theppangna, National Animal Health Laboratory, Ministry of Agriculture and Forestry
• Sengsay Phonthasy, National Animal Health Laboratory, Ministry of Agriculture and Forestry
International partner:
• National Microbiology Laboratory Branch, Canada Funded by Global Affairs Canada
Period of the project: 01 SEP 2021 – 31 AUG 2022
Background
It is estimated that 60% of existing and 75% of emerging infectious diseases have zoonotic origins (Jones, K. et al., 2008). In the past two decades, the world was bombarded with many outbreaks of viruses such as Severe Acute Respiratory Syndrome Corona Virus Infection (SARS- CoV-1), Influenza A H1N1 2009, Middle East Respiratory Syndrome (MERS) CoV, Ebola Virus, and SARS-CoV2 etc. These viruses are believed to have originated in animals and then were transmitted directly or via intermediate host to humans. In addition to the well-characterized human pathogenic and putative viral pathogens, it is postulated that there are certainly other undiscovered zoonotic viruses lurking in animals (Carroll, D. et al., 2018). This emphasizes the importance of continual surveillance and studies of these novel putative zoonotic viral pathogens.
In particular, upstream research that focuses on gaining an understanding of the mechanism of spillover events is crucial to mitigate the risks of future pandemic.
Laos is a landlocked country located at South-East Asia, a region that is recognized as a hotspot for emerging infectious diseases (EIDs) (Jones K.E., et al. 2008). However, Laos still has a weak EIDs surveillance system and limited laboratory capacity for rapid detection of novel emerging viral pathogens. Hence, it is essential to build and strengthen the local capacity for safe investigation and response to emerging zoonotic viral diseases.
In order to build sustainable local capacity in Laos to investigate and respond to emerging zoonotic viral diseases and reduce the threat of potential deliberate use of these novel pathogens, this pilot project builds and fosters scientific cooperation between Canada and Laos, and is co-implemented by the Institut Pasteur du Laos and the National Microbiology Laboratory of the Public Health Agency of Canada.
Objectives
• To build sustainable local capacity in Laos to safely investigate and respond to emerging zoonotic viral diseases
• To establish an inventory and mapping of putative viral pathogens and their animal hosts
• To build and foster scientific cooperation between Canada and Laos
Methodology
Site selection
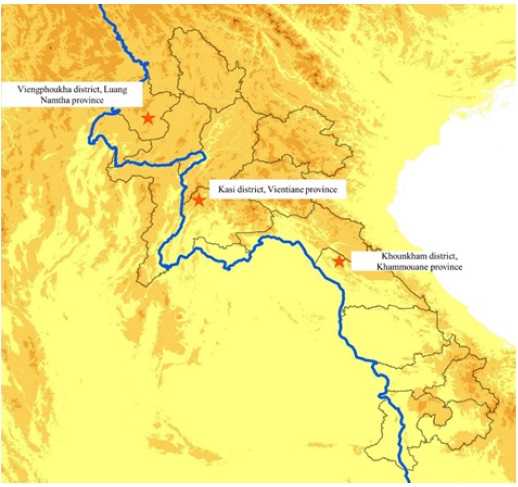
Four field sample collections were conducted in 3 provinces of Laos (Figure 1):
• Field mission 1: Kasi district, Vientiane province (loc: 19.15°, 102.13°) between February 12 and 24, 2022.
• Field mission 2: Viengphoukha district, Luang Namtha province (loc: 20.74°, 101.18°) between March 19 and April 04, 2022.
• Field mission 3: Khounkham district, Khammoune province (loc: 18.16°, 104.43°) between May 19 and 27, 2022.
• Field mission 4: Kasi district, Vientiane province (loc: 19.15°, 102.13°) between June 26 and July 3, 2022.
From these 4 field missions, our team conducted bat capture, rodent capture, and local wet market surveys for sample collections from all wildlife that the vendors who allowed us to examine their wildlife.
Figure 1: Sample collection sites
Field sample collection
Bats
a) Bat collection and identification
Bats were collected using four-bank harp traps (Francis, 1989) and mist nets (Figure 2). Harp traps were set across natural trails and over small streams in the forest understory, and at the entrance of caves in relatively concealed conditions.
Mist nets were also set in similar places to harp traps, but were more often used in open spaces. Both harp traps and mist nets were set before sunset; harp nets were left overnight.
Captured bats were held individually in cloth bags. Species, sex, age (adult or juvenile), and reproductive condition (pregnant or lactating) were determined in the field. Species were identified following Francis (2008), Csorba et al. (2003), and Corbet and Hill (1992). Adults or juveniles were identified by the presence of unfused epiphyses of the phalanges and metacarpal joints (Brunet- Rossinni and Wilkinson, 2009). The reproductive status of female bats was determined by examining the nipples (Racey, 2009). Some external characteristics, including forearm length (FA), were measured using calipers and body mass (W) was taken using a Pesola spring balance. Most bats were marked with wing bands for individual identification and were released at the capture point within 12 hours.
For each species, two or three specimens were retained to confirm identification. Bats were euthanized using chloroform. Specimens were fixed in 95% ethanol in the field and were transferred to 70% ethanol when they were brought back to the laboratory. All specimens were registered and catalogued in the Zoological Collection of the Faculty of Environmental Sciences, National University of Laos, Lao PDR.
Figure 2: Harp trap and mist net placement Harp trap Mist net

b) Bat sampling
Live bats were placed into porous cotton bags (with drawstring mouths) and kept in a cool dry place until sampling time, not exceeding 6 hours. Bats were weighed (in grams) in bags using a Pesola hanging scale or a tabletop scale with or without a container (such as a cup). Once the bat was removed from the bag for sampling, the bag was re-weighed and subtracted from the previous total. Saliva, fecal/anal, urine, blood samples were collected. After sampling, live bats were released at the collection site. Other tissues such as liver, spleen, lung, brain etc. were collected from dead bats.
Rodents
a) Rodent collection and identification
Rodent habitats were identified, especially at entrance of caves and peri karstic areas. Small wire live-traps (14x14x30 cm in WxHxL), Sherman traps, or other similar devices specially fitted for small mammals were set for rodent collection. Between 50 and 100 traps per night were placed in the format of either grid or transect according to the topography, and each trapping session lasted from 6-8 nights. Traps were baited with appropriate baits (e.g., bananas, sticky rice, or dried fish) and checked every morning.
For each species of rodent, two or three specimens were retained to confirm identification. Rodents were euthanized using chloroform. Specimens were fixed in 95% ethanol in the field and were transferred to 70% ethanol when they are brought back to the laboratory. All of specimens were registered and catalogued in the Zoological Collection of the Faculty of Environmental Sciences, National University of Laos, Lao PDR.
b) Rodent sampling
After being euthanized, saliva, fecal/anal, urine, blood, and tissues samples were collected.
Animals from wet market
Dead wild animals such as bats, rodents, pangolins, civet cats, snakes etc. were observed in the local markets. When such animals were found, the local guide borrowed some of them, and brought them to the field station for sample collection. Saliva and fecal/anal samples were collected. Then, those animals would be returned to the vendors.
Laboratory testing
a. Nucleic acid extraction
Total nucleic acids (DNA/RNA) were extracted from bio-samples, initially from bat and rodent anal swabs by using Nuclo Spin®8 Virus (MACHEREY-NAGAL, Germany).
b. Primer design
The degenerate primers targeting the S1/S2 region were designed using HYDEN software (Highly Degenerate Primers: http://acgt.cs.tau.ac.il/hyden/). Briefly, a set of related DNA sequences were aligned, and the degenerate primer pairs were constructed to match all the given sequences with a maximum of three mismatched bases. The detailed algorithm is described in C. Linhart et al. 2002.
c. cDNA synthesis and RT-PCR
cDNA was synthesized using the Maxima H minus first strand cDNA synthesis kit (Thermo Scientific) with random hexamers following the manufacturer’s instructions. Pan-coronavirus RT-PCR assay targeting RNA-dependent RNA Polymerase (RdRP) gene was used for the initial screening of coronaviruses as previously described elsewhere (Chu et al. 2011).
d. Sanger sequencing
PCR products of the expected size were directly sequenced on both strands by Sanger sequencing using the nested PCR primers.
The sequences obtained were confirmed by similarity analysis using the NCBI BLASTn search (http://www. ncbi.nlm.nih.gov/BLAST).
Preliminary results
Field sample collection
From these 4 field missions, our team conducted bat capture, rodent capture, and local wet market surveys from all wildlife that the vendors who allowed us to examine their wildlife.
A total of 1,312 bats (45 species), and 54 rodents (19 species) were captured, and 119 wildlife specimens (21 species) from local markets were examined and sampled during our surveys. More than 4,446 biological samples including anal, saliva, urine, and other tissues were sampled as detail below:
• For bats, 1,208 anal, 1,251 saliva, 474 urine swab and 566 tissue samples were collected (Table 1).
• For rodent, a total of 54 anal, 54 saliva, 27 urine and 228 tissue samples were collected (Table 2).
• For local market, 119 anal, and 119 saliva swabs were collected from wildlife found at the local wet markets (Table 3).
Table 1: Total number of bats captured and biological samples collected.
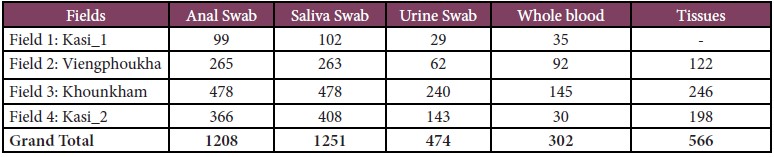
Table 2: Total number of rodents captured and biological samples collected.
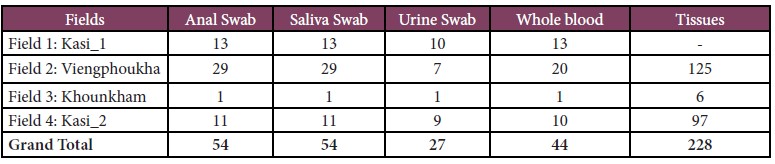
Table 3: Total number of wildlife observed, examined and biological samples collected from wet markets.
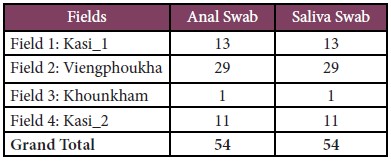
Table 4: Pan-coronavirus screening results.
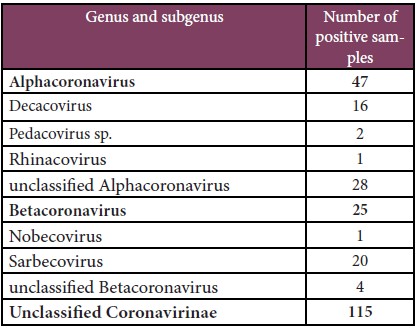
Pan-coronavirus screening
A total of 2,515 extracted samples were screened for coronaviruses at IPL. 187 samples were found to be positive. The NCBI blast results showed 47 samples were Alphacoronavirus, 25 were Betacoronavirus, and 115 were Unclassified coronavirinae. Among Betacoronaviruspositive samples, 20 samples belonged to the subgenus of Sarbecovirus, 4 were unclassified Betacoronavirus, and 1 was Nobecovirus (Table 4).
Phylogenetic analysis
The genomic sequences of partial RdRP of 187 positive samples were used to construct a phylogenetic tree. A high genetic diversity of coronaviruses was detected (Figure 3). Importantly, many sarbecovirus-positive samples were close to both lineages A and B of SARS-COV-2.
Figure 3: Phylogenetic tree
S1/S2 region alignment
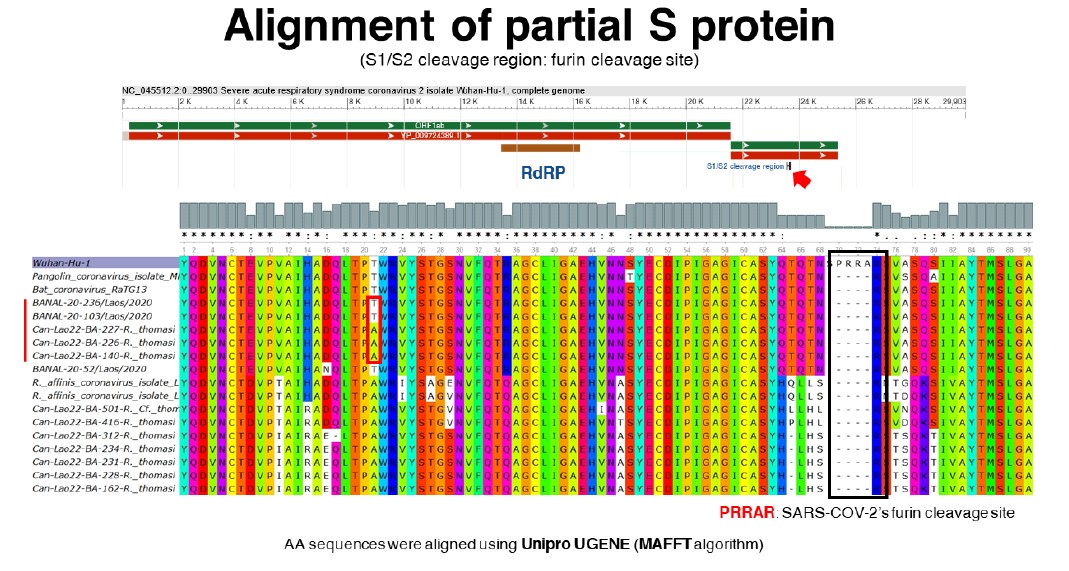
So far, 10 samples of Sarbecovirus were subjected to sanger sequencing of the S1/S2 cleavage region. The phylogenetic analysis of partial RdRP and S protein of those 10 samples was carried out. For the partial RdRP protein, two samples (Can-Lao22-BA-140-R. thomasi and Can-Lao22-BA-226-R. thomasi collected during field 2: Viengphoukha district, Luangnamtha province) were in the same clade as human SARS-COV-2, RaTG13, and Bat coronaviruses from Feung district that were found in Laos in 2020 (Temmam, S. et al., 2022). Furthermore, the phylogenetic analysis on partial S protein placed 3 samples (Can-Lao22-BA-140-R. thomasi, Can-Lao22-BA-226-R. thomasi and Can-Lao22-BA- 227-R. thomasi) close to human SARS-COV-2, RaTG13, and Bat coronaviruses from Feung district (Figure 4). These results were consistent with the alignment of S1/S2 cleavage region showing that 3 samples (Can-Lao22-BA-140-R. thomasi, Can-Lao22-BA-226-R. thomasi and Can-Lao22-BA-227-R. thomasi) shared highly similar amino acid sequences with human SARS-COV-2, RaTG13, and Bat coronaviruses from Feung district (Figure 5).
Figure 4: Phylogenetic trees of partial RdRP (130 Amino acids) and S protein (90 amino acids).
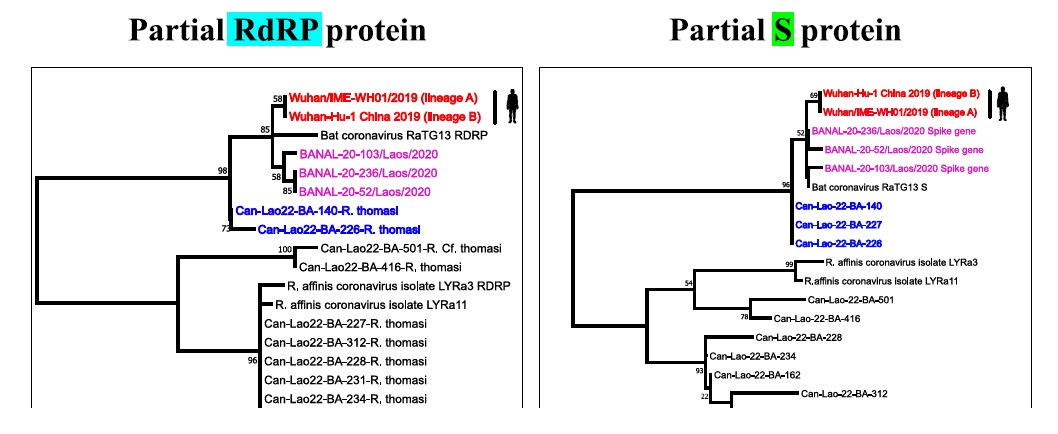
The trees were constructed using Maximum-likelihood method with Jones-Taylor-Thornton (JTT) model and 500 bootstrap replicates in Mega 11 software.
Figure 5: Alignment of S1/S2 cleavage region.
Conclusion and ongoing activities
The results from this pilot phase of the project showed that 187 out of 2,515 samples were positive for coronaviruses. Of these, unclassified coronaviruses yielded the highest number, followed by alphacoronaviruses and betacoronaviruses, respectively. The phylogenetic analysis of the partial RdRP gene implied that the majority of betacoronavirus-positive samples were in the same clade as the lineages A and B of SARS-COV-2. Furthermore, the analysis of S1/S2 cleavage region of 10 Betacoronavirus-positive samples showed that 3 samples (Can-Lao22-BA-140-R. thomasi, Can-Lao22- BA-226-R. thomasi and Can-Lao22-BA-227-R. thomasi) shared highly similar amino acid sequences with human SARS-COV-2, RaTG13, and Bat coronaviruses found in Feung district in Laos in 2020. These results underline the importance for further in-depth investigations such as serological testing, next-generation sequencing, viral characterization etc. to gain insightful understanding of those viruses.
The main focus during the pilot phase was to investigate the presence of coronaviruses at the selected sites, and the initial testing has provided us the preliminary inventory and mapping of the coronaviruses and their animal hosts. Moreover, in the upcoming phase of the project, we are also planning to set up the protocols to screen for other viruses such as filoviruses, paramyxoviruses, rhabdoviruses, arenaviruses etc.
Reference
– Brunet-Rossinni, A.K. and G.S. Wilkinson. 2009. Methods for age estimation and the study of senescence in bats. Pp: 315-325, in Ecological and behavioural methods for the study of bats, 2nd edition (T.H. Kunz and S. Parsons, eds). The Johns Hopkins University Press, Baltimore, xvii + 901 pp.
– Carroll, D., Daszak, P., Wolfe, N. D., Gao, G. F., Morel, C. M., Morzaria, S., … & Mazet, J. A. (2018). The global virome project. Science, 359(6378), 872-874.
– Chu, D. K. W. et al. Avian Coronavirus in Wild Aquatic Birds. Journal of Virology 85, 12815–12820 (2011).
– Corbet, G.B. and J.E. Hill. 1992. The mammals of the Indomalayan region: a systematic review. Natural History Museum Publications and Oxford University Press, 488 pp.
– Csorba, G., P. Ujhelyi, and N. Thomas. 2003. Horseshoe bats of the World (Chiroptera: Rhinolophidae). Alana
Books, xxxii + 160 pp.
– Francis, C.M. 1989. A comparison of mist nets and two designs of harp traps for capturing bats. Journal of Mammalogy, 70: 865-870.
– Francis, C.M. 2008. A field guide to the mammals of Thailand and South-east Asia. New Holland Publishers (UK) Ltd and Asia Books Co., Ltd, Bangkok, Thailand, 392 pp.
– Jones, K. E., Patel, N. G., Levy, M. A., Storeygard, A., Balk, D., Gittleman, J. L., & Daszak, P. (2008). Global trends in emerging infectious diseases. Nature, 451(7181), 990-993.
– Linhart, C., & Shamir, R. (2002). The degenerate primer design problem. Bioinformatics, 18(suppl_1), S172-S181.
– Racey, P.A. 2009. Reproductive assessment of bats. Pp: 249-264, in Ecological and behavioural methods for the study of bats, 2nd edition (T.H. Kunz and S. Parsons, eds). The Johns Hopkins University Press, Baltimore, xvii + 901 pp.
– Smith,C., C. DeJong and H.E. Field (2010) Sampling small quantities of blood from microbats. Acta Chiropterologica. 12(1): 255-258.Bates, P.J.J. and D.L. Harrison. 1997. Bats of the Indian Subcontinent. Harrison Zoological Museum,Sevenoaks, Kent, England, xvi + 258 pp.







