Mapping of Sandfly vectors and their related pathogens in Laos
 Collaborations.
Collaborations.
U.S. Naval Medical Research Unit-2, Singapore
Lao-Oxford University-Mahosot Hospital-Wellcome Trust Research Unit (LOMWRU), Vientiane, Lao PDR
Pathogen Discovery Laboratory, Institut Pasteur, Paris, France.
Fundings.
U.S. Naval Medical Research Center-Asia (NMRC-A)/ Department of Defense Global Emerging Infections Surveillance and Response System (DoD-GEIS) French Ministry of Higher Education and Research (MESRI), France.
Objectives.
The objectives of this project are: 1) to make an inventory of sandfly species that have been collected from different areas in Laos since 2012 and stored at the IP du Laos repository; the taxonomy is based on morphology description and molecular analysis (cytochrome sequencing); 2) to screen the sandflies from IPL depository for phleboviruses by RT-PCR, for other arboviruses by NGS, and for Leishmania spp. by molecular techniques (conventional PCR and sequencing).
Background.
Phlebotomine sandflies are tiny insects encompassing many genera of blood-feeding (hematophagous) flies belonging to the order Diptera, family Psychodidae, and subfamily Phlebotominae. Over the last decades, although the knowledge of sandfly systematics constantly improved and led to improved classification at the species level, the definition of some groups at the genus or subgenus is still under debate 1,2. Most specialists accept 8 genera in the old world: Phlebotomus, Sergentomyia, Grassomyia, Spelaophlebotomus, Spelaeomyia, Parvidens, Idiophlebotomus and Chinius 1. Two of them, Phlebotomus and Sergentomyia, are mostly represented in the Old World and at least 39 species of the genus Phlebotomus are recognized to bite humans 3. Sandflies are principally present in the warm zones of Asia, Africa, Australia, southern and Central Europe and the Americas 4.
Sandflies serve as vectors for several established, emerging and re-emerging infectious diseases such as Carrion’s Disease or bartonellosis, and leishmaniasis. They are also well-known transmitters of viral neuroinvasive infections that may represent significant health problems. Sandfly fever viruses have been reported in Africa, around the Mediterranean basin, in the Middle East and in Central Asia.
Before the 1990s, the South-East Asia (SE Asia) region, was considered as a region free of autochthonous transmission of leishmaniasis and other sandfly related diseases. Thus, this lack of evidence of any involvement of phlebotomine sandflies in disease transmission dramatically limited studies on sandflies. Only some work on the systematic of sandflies was conducted in the region from the 1930s up to the early 1980s 5-8. However, interest in Phlebotomine sandflies from this area increased since the first autochthonous human case Sandfly-borne viruses are unknown in SE Asia. There is a single serological study seeking the presence of phleboviruses in SE Asia in which the authors could not find any evidence for the circulation of phleboviruses in the region 10. However, this study used the phlebovirus strains that circulated elsewhere which may differ from local strains.
In Laos, data on the bionomics of sandflies and sandfly-borne pathogens are scarce. To our knowledge, very few studies on sandflies have been performed in the country; two species of Phlebotomus and five species of Sergentomyia were initially reported 7 and more recently one species of Chinius was described from the country 11.
Here we propose to make an inventory of sandfly species and sandfly-borne pathogens in Laos by investigating sandfly samples in the Institut Pasteur du Laos repository. More than 10,000 female and male sandflies were stored in IP-Laos -80⁰ freezers. This is the biggest known sandfly collection in Laos. These sandflies were collected by different projects of IP du Laos in different areas of Laos from 2012 to the present.
Methodology.
Working group.
In order to identify common and emerging vector-borne diseases including viral and leishmania pathogens in Laos, this study was conducted during 2022-2023 in collaboration with the Pathogen Discovery Laboratory at Institut Pasteur Paris, and the Lao-Oxford University-Mahosot Hospital-Wellcome Trust Research Unit (LOMWRU), in Vientiane (Fig. 1).
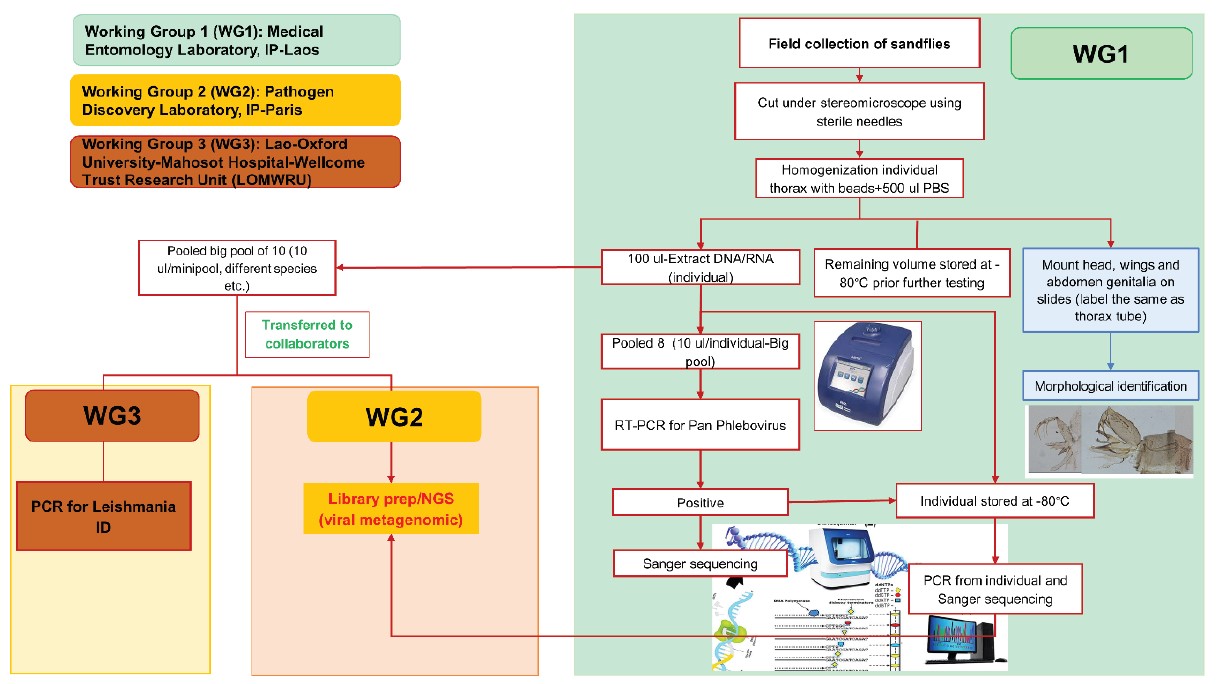
Figure 1: Working groups and workflow of this project.
For 645 samples, sandfly bodies were homogenized, and nucleic acids were extracted individually without cutting and morphological identification. Only sex was identified. A molecular technique was used for species identification by comparing the results with our cyt-b database as described above.
Total nucleic acid extraction.
Individual thorax/whole body in a NucleoSpin® 8 tube was filled with 500 μl of Phosphate-Buffered Saline (PBS) 1X and Lysing Matrix E beads or A (MP Biomedicals). A TissueLyser II system (Qiagen) was used for homogenization for 10 min at a vibration frequency of 25/s. To obtain total nucleic acids extraction (DNA/ RNA) for pathogen screening, a total of 100 μl of each tube was extracted using a NucleoSpin® 8 Virus extraction kit following the manufacturer’s protocol. The remaining 400 μl of each pool was stored at –80°C for further pathogen isolation.
Total nucleic acids extraction (DNA/RNA) from each sample was then pooled from 5-10 individuals by transferring 10 μl of each sample to a pool. Aliquoted samples were then used for viral screening and sent to Lao- Oxford- Mahosot Hospital- Wellcome Trust Research Unit (LOMWRU) for Leishmania spp. detection as described below.
Reverse transcriptase polymerase chain reaction for virus screening (IPL).
Primers for pan-genus phlebovirus were selected and tested for screening sandfly specimens for the presence of phlebovirus sequences by means of conventional nested RT-PCR as previously described by Sanchez- Seco, et al., 2003 14. Once a pool was found positive, the corresponding individual samples were retested to find the positive individual.
Next generation Sequencing (IP Paris).
Extracted products from positive samples were sent to IP Paris for NGS analysis. In brief, at the Pathogen Discovery lab (IP-Paris), the quality and concentration of samples were evaluated using an Agilent 2100 Bioanalyzer and Qubit 2.0 Fluorometer (Invitrogen, USA) respectively. Total RNA libraries were constructed using the SMARTer Stranded Total RNA-seq Kit v3-Pico input mammalian (TaKaRa). DNA library quality and concentration were evaluated using the same techniques mentioned above. RNA sequencing was performed on the Illumina NextSeq2000 sequencer, in pairedend 2 x101 pb format (to achieve approximately 50 million reads), at the Biomics platform (Institut Pasteur, Paris). Bioinformatics analyses were carried out using a bioinformatics pipeline (Microseek) developed by the pathogen discovery lab team (see https://pubmed.ncbi. nlm.nih.gov/36146797/).
Leishmania screening at LOWMRU.
PCR was performed on pooled DNA extracted from sandflies at the Molecular Department at LOMWRU based on the method previously described by Shonian, et al., 2003 15 using primers LITSR and L5.8S with a PCR product of 300-350 bp.
Results.
Sandfly sample used in this study and species composition.
A total of 3,121 sandflies collected from 8 districts from 5 provinces were prepared for pathogen screening (Table 3). Total nucleic acid (DNA/RNA) was extracted from 2,524 sandflies and the specimens were screened for phlebovirus detection. A total of 1,772 samples were prepared by cutting the head and genitalia for morphological identification, and only the thorax was used for phlebovirus screening; 645 samples were screened without morphological identification; and 107 samples remain to be identified.
From a total of 1,772 samples, sandflies were morphologically classified into 26 species of 5 genera.
Phlebovirus screening at IPL.
More detailed results will be provided in an upcoming publication.
Samples testing by NGS at IP Paris.
A total of 24 samples were used for NGS analysis: 9 big pools (including 2 negative pools), and 15 individuals.
More detailed results will be provided in an upcoming publication.
Leishmania screening.
More detailed results will be provided in an upcoming publication.
Discussion.
This is the first largest inventory of sandflies in Laos. At least 26 species with 13 new records species have been updated to the list of sandfly fauna of Laos.
References.
1. Akhoundi M, Kuhls K, Cannet A, Votypka J, Marty P, Delaunay P, et al. A Historical Overview of the Classification, Evolution, and Dispersion of Leishmania Parasites and Sandflies. PLoS neglected tropical diseases. 2016;10(3):e0004349. 2. Rispail P, Leger N. Numerical taxonomy of Old World Phlebotominae (Diptera: Psychodidae).
2. Restatement of classification upon subgeneric morphological characters. Memorias do Instituto Oswaldo Cruz. 1998;93(6):787- 93.
3. Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Medical and veterinary entomology. 1990;4(1):1-24.
4. Killick-Kendrick R. The biology and control of phlebotomine sand flies. Clinics in dermatology. 1999;17(3):279-89.
5. Lewis DJ. The phlebotomine sandflies Diptera: Psychodidae of the Oriental Region. Bulletin of the British Museum (Natural History). 1978;37:217-343.
6. Lewis DJ. A Taxonomic Review of the Genus Phlebotomus (Diptera, Psychodidae). British Museum (Natural History). 1982.
7. Quate LW. A review of the indo-chinese phlebotominae. Pacific insects. 1962;4(2):251-67.
8. Raynal J. Contribution à l’étude des phlébotomes d’Indochine. Généralites Arch Inst Pateur Indochine. 1935;19:337-69.
9. Thisyakorn U, Jongwutiwes S, Vanichsetakul P, Lertsapcharoen P. Visceral leishmaniasis: the first indigenous case report in Thailand. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1999;93(1):23-4.
10. Tesh RB, Saidi S, Gajdamovic SJ, Rodhain F, Vesenjak- Hirjan J. Serological studies on the epidemiology of sandfly fever in the Old World. Bulletin of the World Health Organization. 1976;54(6):663-74.
11. Leger N, Depaquit J, Gay F. Chinius eunicegalatiae n. sp. (Diptera; Psychodidae), a cavernicolous sandfly from Laos. Annals of tropical medicine and parasitology. 2010;104(7):595-600.
12. Leng, Y. J. (1987). A preliminary survey of phlebotomine sandflies in limestone caves of Sichuan and Guizhou Provinces, south-west China, and description and discussion of a primitive new genus Chinius. Ann Trop Med Parasitol 81(3): 311–317.
13. Leger, N., J. Depaquit and F. Gay (2010). Chinius eunicegalatiae n. sp. (Diptera; Psychodidae), a cavernicolous sandfly from Laos. Ann Trop Med Parasitol 104(7): 595–600.
14. Sanchez-Seco, M.P., et al., Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J Med Virol, 2003. 71(1): p. 140-9.
15. Schönian et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagnostic Microbiology and Infectious Disease, 2003; 47: 349-358 Hematophagous insects and ectoparasites in karstic cave areas and their related pathogens in Laos Collaboration Lao Army Institute for Disease Prevention Young






