Arbovirus ecology
Project coordinator: Kouxiong XAYTENG Member of
staff: Souksakhone VIENTPHOUTHONG

Discussing the dengue virus outbreak with deputy of Xaythany district and head of Oudomphon village
Exploration studies were held to continue activities that provided pertinent preliminary data on ticks and sandflies or to open new fields of investigation.
Tick Map 1.5
Research program funded by the US DOD In collaboration with NMRC-A, Singapore.
Vector-borne diseases constitute a significant infectious disease risk for deployed military personnel and for local populations. In Laos, definitive diagnosis is often not available for vector-borne illnesses, so the infectious diseases that are a threat to military and civilian populations are not well defined. Tick characterization and investigations for the presence of pathogens already provided evidence of their important role in disease transmission in the Lao PDR.
Our first investigation of Laotian ticks and tick-borne pathogens was launched in late 2012 and continued into 2014 through collaboration between the U.S. Naval Medical Research Center-Asia (NMRC-A) and the Institut Pasteur du Laos (IP-Laos). Its goals were to determine the geographical distribution of putative tick vectors of bacterial and arboviral diseases in the Nakai Nam Theun National Protected Area (NNT NPA), known as the Watershed Management and Protection Authority area (WMPA), located in Nakai District, Khammuan Province, Laos. The results showed that five genera comprising at least eleven species (including three suspected species that could not be readily determined) were identified from 215 adult specimens. These collections, together with the literature to date, provide evidence for the occurrence of at least twenty-two ixodid tick species, representing six genera. At least four tick species were newly recorded from the WMPA area, Nakai District, Khammuan Province (Vongphayloth, Brey et al. 2016).
A new collection campaign has been designed to increase the number of adult tick specimens to facilitate species identification. Table 1 summarizes the results of tick specimens for arbovirus screening.
Table 1. Arbovirus screening by RT-PCR targeting flaviviruses, alphaviruses, and phleboviruses from tick samples
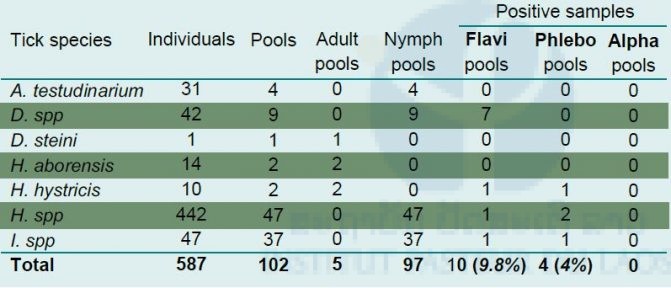
Flavivirus sequences were evidenced in engorged Dermacentor spp pools (n=4) and Ixodes spp (n=1) pools. Engorged ticks were retrieved from two orders of small mammalians (Rodentia; Scandentia). Flavivirus sequences could also be detected in pools of non-engorged Dermacentor spp (n=3), Haemaphysalis spp (n=1) and Haemaphysalis hystricis (n=1) pools. Phleboviruses were found only in non-engorged tick pools (Table 1). The co-detection of flavivirus and phlebovirus sequences was found within pools of Haemaphysalis spp (n=1) and Haemaphysalis hystricis (n=1).
Results of RT-PCR screening were used to select candidate samples for virus isolation assays. All tissue supernatants from pools found positive for flavivirus were inoculated on C6/36 and VeroE6, whereas phlebovirus positives were only inoculated on VeroE6 cell lines. Of the ten isolation assays from flavivirus-positive samples, three gave a positive result. Of these, only two could be maintained for several passages. Within the four samples found positive for pan-phlebovirus RT-PCR, two led to a positive culture after inoculation on Vero E6 cells. Both could be maintained over three consecutive passages. RT-PCR products for the culture screening were purified to be submitted to Sanger sequencing but this approach failed to provide relevant data for viral species identification. Efforts now focus on amplification of the first passage isolates to verify their stability in culture and obtain sufficient material to attempt a deep-sequencing approach for a full viral identification and phenotypic characterization.
The present project was set up to confirm preliminary data on tick species identification and pathogen screening obtained for the first time in the Lao PDR (see TickMap reports; Taylor, 2016). The collection strategy was modified as most of the tick specimens collected in the previous study were nymphs. Direct collection of wild or domestic animals allowed for obtaining some adult specimens. Even if the total number of adults remains low, this approach is helpful to determine the trophic preferences of the species.
Bio-Lao
Research program funded by the US DOD In collaboration with NMRC-A, Singapore.
The Bio-Lao project aims to initiate practical actions to expand knowledge of vector-borne infections in the Lao PDR. The proposed approach combines training sessions and research activities in two provinces. These mixed activities, involving the provision of concrete materials and events, include demonstrating and evaluating diagnosis, surveillance, and control strategies.
Some members of the three main arbovirus families involved in human diseases (i.e. Flaviviridae, Togaviridae and Bunyaviridae) have Culex mosquito species as major vectors. However, available data on these “Culex-borne viruses” in South-East Asia are poor, except for Japanese encephalitis.
A total of 1076 adult Culex female mosquitoes of nine species (Table 2; refer to Medical entomology unit section) was divided into two groups i.e. non-engorged and engorged mosquitoes. The non-engorged specimens were dissected: wings and legs were pooled (maximum ten per pool), while the thoraxes and heads were individually placed in test tubes. The abdomens and head/thoraxes of engorged mosquitoes were put transfered in two independant tubes. The 319 samples which were obtained (112 non-engorged pools and 207 engorged mosquitoes) were screened for flavivirus, alphavirus, and phlebovirus. sequences
Table 2. Detection of flavivirus and alphavirus genomic sequences by RT-PCR in Culex specimens
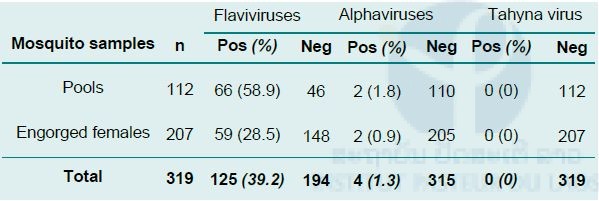
None of the samples tested were found positive for the presence of phlebovirus sequences.
A positive result for flavivirus sequences detection was obtained for 58.9% of the pools (Table 2). As shown in Table 3, most of the positive pools corresponded to mono-specific pools of Culex vishnui (25/39; 64%). Of the 207 engorged Culex females, 59 (28.5%) were found positive for flavivirus sequences. Most of them were found within Culex vishnui caught in the two provinces. Of the seven clearly identified Culex species, five were detected positive for flavivirus sequences (Table 3).
Table 3. Flaviviruses and alphaviruses detected in engorged Culex females
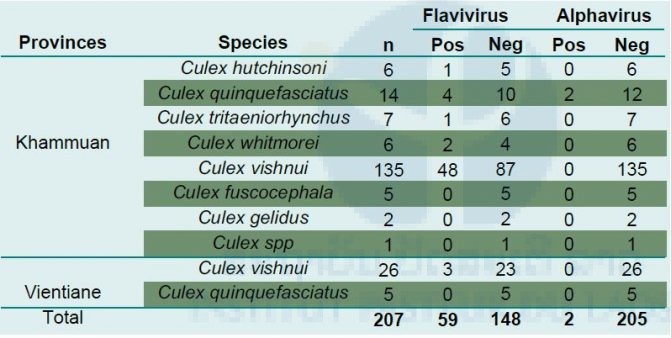
Results of RT-PCR screening were used to select candidate samples for virus isolation assays. All tissue supernatant from pools and individual specimens found positive for flavivirus and alphavirus sequences were inoculated on a C6/36 cell line. Among the 156 isolation assays, a total of 28 culture supernatants gave a positive signal for flavivirus sequences. Of these cultures, all 28 were obtained from specimens collected in Khammuan province. Most of the isolates were obtained from C. vishnui tissues (n=24), two from C. quinquefasciatus and one each respectively from C. hutchinsoni and C. whitmorei. RT-PCR products for the culture screenings were purified and submitted to Sanger sequencing to attempt identifying the viral species. Preliminary results didn’t allow sufficiently accurate data to draw any partial conclusions as most signals were of low quality. Efforts now focus on amplification of the first passage isolates to verify their stability in culture and obtain sufficient material to attempt a deep sequencing approach for viral identification and phenotypic characterization.
Tentative assays to isolate alphaviruses from pools (n=2) and head–thorax segments (n=2) were unsuccessful. A new approach using “micro-culture” on culture slides is ongoing to increase chances of isolating arboviruses from tissues with low viral loads.
Investigation of Zika virus infection in mosquitoes in Khammuan Province and Vientiane Capital
Research program funded by the US DOD In collaboration with NMRC-A, Singapore.
Background:
Zika virus was first isolated in Uganda in 1947 from a monkey in the Zika forest. A strain of Zika virus was isolated in 1969 from a pool of Aedes aegypti mosquitoes in West Central Malaysia. Serologic evidence of human infection by Zika virus has been observed in Malaysia, Thailand, Vietnam, and the Philippines since 1960. Surprisingly, a retrospective study conducted by the arboviruses and emerging viral diseases laboratory, IPL, indicated that Zika virus infection and Zika–dengue viruses co-infection have been detected from suspected dengue cases collected since 2012–2013 when the Lao PDR experienced a huge dengue epidemic.
Objectives:
To document Zika virus maintenance and transmission cycles in the Lao PDR.
Method:
A retrospective study of Zika virus infection in mosquitoes collected at the locations of dengue suspected and confirmed cases in Vientiane Capital in 2013 during the dengue outbreak period and in Khammuan Province in 2012. The samples included adults and larvae of female and male Aedes aegypti mosquitoes and all larvae were reared to adult stage.
Results:
A total of 113 Aedes mosquito samples (89 pools of adults emerged from larvae; 16 pools of adults collected in Vientiane Capital and 8 pools of adults collected in Khammuan Province) were screened for the presence of Zika virus sequence by a real time RT-PCR technique. The results showed that no Zika virus was detected from the mosquito samples analyzed. A larger sample size is required.
Investigation of Tahyna virus in bats and rodents in Vientiane and Khammuan Provinces
Research program funded Institut Pasteur du Laos.
Background:
Tahyna virus (TAHv) was first isolated from mosquitoes in Czechoslovakia in 1958. Human diseases associated with TAHv have been reported since the 1960s. Recently, TAHv has been isolated from mosquitoes collected in China in 2006. TAHv-specific antibodies have been detected in several nonhuman mammals. In Laos, a preliminary seroprevalence study evidenced anti-TAHv IgG in dengue-suspected cases collected in Vientiane Capital in 2016 and in Khammuan Province in 2013. Up to now, no TAHv has been detected among these cases. However, vectors and hosts are abundant in the region, raising the question of possible maintenance and transmission cycles of TAHv.
In 2015, bats and rodents were captured in Vientiane Province and in National Protected Areas of Khammuan Province. The specimens were identified for species and blood samples were taken and transferred to IPL Lab. In this study, samples were randomly selected within the series of the different collection sites and mammal species and tested for the TAHv virus genome.
A total of 116 samples, including 100 bat samples of 21 species collected in 7 sites and 16 rodent samples of 7 species collected in 3 sites, were tested for TAHv by real time RT-PCR but no TAHv sequence could be found in these series.
Screening of human samples and mosquito specimens is ongoing to attempt to obtain local isolates of TAHv.
- Picture 1. Blood collection in bats
- Picture 2. Blood collection from rodents