Arbovirus ecology
Project leader: Dr. Kouxiong Sayteng
Member of staff: Souksakhone Viengphouthong
Background
Two main project in the domain of arbovirus cycle description in Lao PDR were achieved in 2015. Both of them were conducted in close collaboration with the medical entomology unit of the Institut Pasteur du Laos.
SandflyMap
Sandflies are known as vectors of viral, protozoan, and bacterial diseases. The sandfly-borne viruses comprise three genera: (1) Phlebovirus (Family Bunyaviridae), (2) Vesiculovirus (Family Rhabdoviridae), and (3) Orbivirus (Family Reoviridae). The most important groups are the Phlebovirus genus, which includes the sandfly fever Sicilian and Toscana viruses, and the Vesiculovirus genus, which includes vesicular stomatitis as well as the Chandipura and Isfahan viruses. The vast majority of sandfly-borne viruses cause only moderately severe diseases in only specific geographical regions, so they are given little attention by physicians and their prevalence is underestimated. The high morbidity is usually among non-native populations such as military personnel and travelers in the endemic regions. A recent review has indicated that sandfly-borne virus diversity in the Mediterranean basin is higher than initially suspected (Maroli, Feliciangeli et al. 2013). Furthermore, recent investigations have shown that flavivirus RNA closely related to insect-only flaviviruses were detected in sandflies (Moureau, Ninove et al. 2010, Sanchez-Seco, Vazquez et al. 2010). There are high possibilities that sandfly-borne viruses may be circulating in Lao PDR as well as in Southeast Asia, but we lack evidence at present because of their self-limited symptoms within a specific area, the absence of a surveillance system, limited laboratory resources for differential diagnosis and a paucity of scientific interest in sandfly-transmitted viral diseases.
The SAND-Map project involved U.S. Naval Medical Research Center-Asia (NMRC-A) and the Institut Pasteur du Laos (IP-Laos). Its goals were to establish an inventory of sandfly species and their distribution in Laos and to assess their role in pathogen transmission, especially for Phleboviruses.
Five sites were selected for sandfly collection during our study—one in Fueng district, Vientiane province; two in Nakai district, Khammuan province and Korbong village, in Watershed Management and Protection Authority area (WMPA) one in Boualapha district, Khammuan province (Phou Hin Nam Nor National Protected Area (PHNN NPA) and one site in Vientiane capital. From May to June 2015, sandfly collections were performed during the field missions of a parallel project (ResArbo-Coll/ResArbopath see below).
Sampling and identification of sandflies have been performed by the entomology team (refer to the medical entomology annual report).
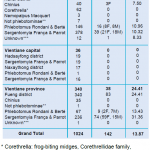
Pan-phlebovirus RT-PCR screening allowed the identification of viral sequences in 142 of the 1,024 (13.87%) sandfly specimens tested so far. Among them, 59/648 (9.1%) and 83/340 (24.41%) of the positive samples were captured, mainly inside caves, in Khammuan province and Vientiane province respectively. No positive samples were found in Vientiane capital. Almost all positive samples were detected within the Sergentomyia and Phlebotomus genera and at a lower level in the Chinius genus. In the genera Phlebotomus and Sergentomyia, both female and male were found positive. In Khammuan province, the positive rates of genus Phlebotomus and genus Sergentomyia were equal at 10% (10.96% and 10.32% respectively). In Vientiane province, the positive rate was 31.36% in the Sergentomyia genus and 13.43% in the Phlebotomus genus (Table 1).
Table 1: Phlebovirus detection by conventional nested RT-PRC.
Most of the samples positive for phlebovirus sequences were found in specimens collected inside caves from Vientiane province (83/142; 58.45%) and from Khammuan province (59/142 ; 41.55%) whereas no samples from Vientiane capital were found positive. Both females and males within the genera Phlebotomus and Sergentomyia were positive for viral sequences. Interestingly, in most cases, high infestation rates were found in males nearly equal to those recorded in females. These results suggest that multiple modes of viral transmission are involved in at least these two sandfly genera (i) direct blood feeding for females, (ii) horizontal sexual transmission between females and males, and (iii) vertical transmission to offspring for both males and females. This is the first time that phlebovirus sequences have been detected in sandflies from Lao PDR. Viral species identification is now our main focus. This will be tentatively achieved by viral isolation assays and genomic sequence analysis.
ResArbo
Arboviruses pose major health problems in Southeast Asia and in both rural and urban areas. Intriguingly, sylvatic transmission (i.e. transmission in the wild environment) has been poorly studied in Asia, including for major pathogens such as dengue and chikungunya. Arbovirus transmission often relies on mosquitoes, especially Aedes and Culex. Distribution of the primary vectors of dengue and Chikungunya (i.e. Aedes aegypti; Ae. albopictus) and of Japanese encephalitis (i.e. Culex tritaeniorynchus) is well documented in Southeast Asia.
However, other groups of arthropods have been recognized as primary vectors of arboviral diseases. Ticks are responsible for the transmission of the Tick-Borne Encephalitis virus (TBE) and have been recently recognized as a vector of the newly discovered Severe Fever and Thrombopenia virus. Sandflies are well known vectors of Leishmania spp parasites but also of Bluetongue virus (Reoviridae) and neuro-invasive phleboviruses (Toscana; Sandfly Naples). Studies of ticks and sandflies have been neglected in Southeast Asia countries.
- Picture n°3: Sample preparation during a Resarbo-Coll project field mission (photo IP Laos)
- Picture n°4: Blood sampling on lived vertebrates during Resarbo-Coll project field mission (photo IP Laos)
These gaps in the inventories of both vectors and amplifying/reservoir vertebrates involved in known and unknown arboviruses may have serious consequences for diagnosis and patient management, epidemiology, and the prevention of human and animal arboviral diseases. Southeast Asia has been considered for decades as an endemic area for dengue, chikungunya, Japanese encephalitis, and numbers of other arboviruses but major questions are still to be answered: i) How do arboviruses maintain themselves in the environment? ii) How do they survive during adverse periods when vectors are not prevalent? Some preliminary data have suggested that animal reservoirs and/or the arthropods themselves could serve as reservoirs. If arboviruses circulate specifically in the wildlife population, the sylvatic cycles could probably switch into the human population in rural areas, then in urban. In order to address these questions, Institut Pasteur du Laos, the National University of Laos, and the Naval Medical Research Center–Asia (NMRC–A) in Singapore proposed a study to assess the temporal–spatial distribution and infection status of arbovirus hosts/vectors and an analysis of putative reservoirs/vectors in karstic and peri-karstic areas of two provinces in Lao PDR
- Picture n°5: Sandflies collection in a cave during a Resarbo-Coll project field mission (photo IP Laos
One of the main objectives of the project was to set up a network of experts in order to collect a large spectrum of animal species (including bats, rodents, amphibians, reptiles, hematophagous arthropods, ectoparasites, etc.) that may play a role in the transmission and/or the maintenance of arboviruses in wild environments. Lao limestone karstic areas, drilled by numerous caves, offer ideal study sites to identify such interactions. ResArbo-Coll was coordinated by the Arbovirus ad emerging viral diseases laborartory. This phase of the ResArbo project focused on the collection and identification of the animal species, will provide the foundations of the ResArbo-Path project that aims to evidence arbovirus markers in the biological material and to identify transmission pathways between species.
ResArbo-Path
Vector-borne diseases constitute a significant infectious disease risk for all populations in Laos. However, in Laos, definitive diagnosis is often not available for vector-borne illnesses, so the infectious diseases that are a threat to the population are often not well defined. One of the major biological questions to address is how do vector-borne diseases maintain themselves in nature? How do they survive during adverse periods when vectors are not prevalent? Some preliminary data have suggested that animal reservoirs and/or the arthropods themselves could serve as reservoirs. In order to address these questions and to identify the putative reservoirs of these known and emerging vector-borne pathogens in Laos, Institut Pasteur Laos, the National University of Laos, and the Naval Medical Research Center-Asia (NMRC-A) in Singapore proposed a study to assess the temporal-spatial distribution and infection status of arbovirus hosts/vectors and to analyze putative reservoirs/vectors.
Resarbo-Path constituted the second phase of the ResArbo project and focused on the screening of the samples collected during phase 1 for a selected panel of pathogens.
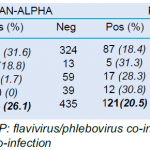
Blood samples from different vertebrate species were investigated by pan-genus (i.e. alphavirus, flavivirus, and phlebovirus) conventional RT-nested PCR. Among the samples available, whole blood and/or dry blood spots, only whole blood samples were analyzed in order to draw a correlation between RT-PCR screening and viral culture assays. A total of 589 blood samples collected from bats, rodents, reptiles and amphibians were tested with the different RT-PCR systems (Table 2). Of them, 385 (65.4%) were found positive for at least one of the targeted viral gerera Respective proportions for alphavirus, phlebovirus, and flavivirus were 26.2% (154/589), 20.5% (121/589) and 18.7% (110/589) respectively (Table 2).
Amphibians and reptiles presented the highest flavivirus sequence positivity rates with 28.2% (11/39) and 26.7% (16/60) respectively, followed by bats with 17.5% (83/474). Alphavirus sequences were found predominantly in 31.7% of bats (150/474) but also in 18.8% (3/16) of rodents and 1.7% /60) of reptiles. All four groups of hosts displayed phlebovirus sequences at significant levels with 18.4% (87/474), 31.3% (5/16), 28.3% (17/60) and 30.8% (12/39) in bats, rodents, reptiles, and amphibians respectively.
- Table 2. Arbovirus screening of vertebrate blood samples by RT-PCR
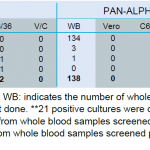
In order to demonstrate actual infection of vertebrates, material from whole blood samples was saved at the beginning of the extraction procedure for immediate inoculation on C6/36 and Vero E6 cell lines. In all, 589 whole blood samples were screened and inoculated on both cell lines.
At this stage, the screening of culture supernatants has been achieved for 452 of the 589 cultures. The 137 remaining passage 1 cultures were stored frozen at −80 °C until RT-PCR analysis. Screening of passage 1 culture supernatants by RT-PCR allowed the detection of flavivirus sequences in 15.9% (72/452) of Vero E6 cultures but from only 0.4% (2/452) of C6/36 culture. Phleboviruses were successfully isolated from 42% (190/452) of the Vero E6 cultures. As shown in Table 4, the number of positive cultures exceeded the number of whole blood samples detected positive for phlebovirus sequences. No alphavirus could be retrieved from either Vero E6 or C6/36 cultures (Table 3).
These results demonstrated that some of the vertebrate species harbor actual arboviral infections. Thus, detection of flavivirus and phlebovirus sequences by RT-PCR on whole blood was not an artifact
- Table 3. Screening of vertebrates samples cultures
At the end of the program, 384 (9%) specimens of the 4232 sandflies could be tested for the presence of phlebovirus sequences because of the time taken for the specimens identification and preparation processes. Among these samples, 11 (2.9%) were found positive. The 11 positive samples were distributed respectively in 4 (36.4%) and 7 (63.6%) of sandflies belonging to the Phlebotomus (ref: Rondani & Berté) and Sergentomyia (ref: França & Parrot) genera (Table 4). Most of the positive specimens were found in females but, in both genera, infected males were also found.
- Table 4: Phleboviruses detected in sandfly genera
Virologic investigation of vertebrate and arthropod specimens collected during two field missions in Vientiane and Khammuan Provinces provided information on arbovirus geographic distribution and possible inter-species relations that may contribute to arbovirus maintenance in wild environments.
Phlebovirus sequences were evidenced in all groups of vertebrates investigated, which comprised bats, rodents, reptiles, and amphibians. Moreover, the presence at the same time and in the same areas of the main recognized arthropod vector of phleboviruses, i.e. sandflies, strongly supports a local transmission of phleboviruses. Interestingly, the presence of viral sequences in males sheds light on possible complex mechanisms of phlebovirus maintenance that may include vertical and horizontal transmission in sandfly populations. The role of the different vertebrate species in this(ese) cycle(s) remains to be determined. Indeed, no conclusion could be drawn at this stage on the significance of the viral sequences detected to determine if vertebrates participate as a reservoir, amplifying host, or dead-end host. However, successful viral isolations from bats, rodents, and reptiles demonstrate that a large spectrum of vertebrates is supporting phlebovirus replication. Sequence data are now needed to identify the viral species and establish the links between arthropods and vertebrates.
Most members within the flavivirus and alphavirus genera are known to be transmitted by mosquitoes. The arthropod collection strategy (type of trap, collection sites) was more adapted to sandflies. This explains, at least in part, the lack of mosquito samples. Nevertheless, evidence of the presence of flavivirus and alphavirus has been established by sequence detection and/or viral isolation.
Flavivirus infection occurred with bats, reptiles, and amphibians but was not evidenced in rodents. Alphavirus detection was also positive in all vertebrate groups except amphibians. However, as the number of specimens was limited, these results do not exclude a possible infection of rodents by flavivirus and amphibians by alphavirus.
Altogether, this exhaustive inventory of the caves’ fauna allows us to speculate about a specific transmission cycle of phleboviruses in the caves. The absence of mosquitoes inside the caves supports the hypothesis of more complex cycles for flavivirus and alphavirus, within which transmission of viruses to cave-dwelling vertebrates occurs during animal displacement outside the caves. For instance, Rattus rattus is a rodent species which often shares human habitats in the villages located near the caves where environmental conditions are more adapted to the proliferation of mosquito species. This does not exclude the existence of alternative vectors for flavivirus and alphavirus. Indeed, recent studies evidenced flavivirus sequences in sandflies and our team found alphavirus sequences in ticks (refer to TickMap project report). Future investigations planned on ectoparasites collected from vertebrates may bring some answers.
Potential dual and triple infections involving the arboviral genera tested have been evidenced in different vertebrates. Among the bat specimens, 3.2% were co-infected by flavivirus and alphavirus, 14.6% were positive for alphavirus and phlebovirus sequences and 1.5% displayed sequences of the three viral genera. This phenomenon was not limited to bats. Even if the specimen number was limited for rodents and reptiles, different patterns of multiple infections could be evidenced. These last results suggest that the vectors transmitting the viruses may have a wide feeding pattern. Sequence analysis will help in exploring this question.
Viral sequences belonging to the three genera were detected in significantly distant study sites. These data support a high viral biodiversity in each province and possibly at the country level. It is not yet known whether the members of the different viral genera present in the two provinces are identical, represent genetic variants of the same species, or are independent species. An extended sequencing program is now needed to establish this degree of biodiversity and to draw up hypotheses on the distribution of the viruses.
This one-year program allowed the gathering of preliminary results on viral biodiversity in two provinces of the Lao PDR. The different sites can be considered as hot spots in terms of rates of infection, diversity of infected vertebrates, and diversity of viral genera circulating at the same time in the same place. As only a unique field mission could be organized for each site, it is not possible to determine the influence of seasonality. However, as the missions took place at the end of the dry season, it can be expected that infection rates and both vertebrate and arthropod diversity may increase temporarily during the rainy season. Cave environments explored during this project offer optimal conditions for virus spillover as illustrated by the infection rates in animal populations. Our study sites in wild environments but located at the limit of human-occupied areas could be used as sentinel sites to assess the risk of viral emergence in human populations.