COVID-19 surveillance in Laos
2 Project Coordinator:
Project Coordinator:
• From October 2021 to May 2022: Dr. Somphavanh
Somlor• From June to October 2022: Dr Cécile Troupin
Staff members:
Dr. Thonglakhone Xaybounsou
Longthor Vachouaxiong
Kedkeo Intavong
Funding: Supported by the Gouvernement du Grand-Duché du Luxembourg; the Agence Française du Développement (AFD); the Foreign, Commonwealth and Development Office of United Kingdom; the Japan International Cooperation Agency (JICA); the Swiss Agency for Development and Cooperation (SDC); the United Nations Resident Coordinator Office and the French Embassy in Laos.
Background
After the emergence of the Severe Acute Respiratory Syndrome – Coronavirus 2 (SARS-CoV-2) virus in the city of Wuhan in China at the end of 2019, which led to the COVID-19 (Coronavirus Disease 19) pandemic, the Institut Pasteur du Laos (IPL) was requested by the Ministry of Health (MOH) to participate in the national response to the pandemic. As consequence, since March 2020 IPL has been one of the frontline laboratories for the diagnosis of SARS-CoV-2 in the country.
RT-PCR testing of SARS-CoV-2
From October 2021 to September 2022, 44 389 samples have been tested at IPL for COVID-19 diagnosis and SARS-COV-2 has been found in 11 954 of them (26.93%). Samples were mostly nasopharyngeal swabs with occasional naso + oropharyngeal swabs.
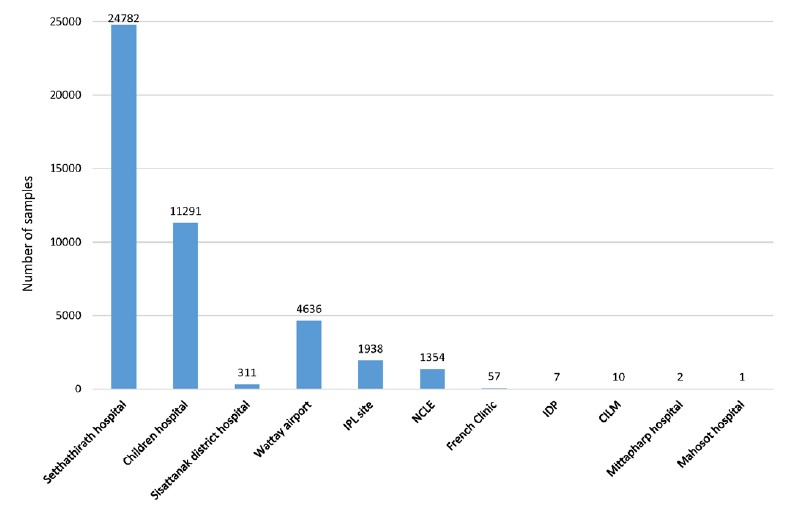
The samples tested at IPL all come from Vientiane capital and are collected through a network that was set-up after discussion with MOH and NCLE, including: i) 3 hospitals in Vientiane capital (Setthathirah hospital, Children hospital, Sisattanak district hospital); ii) Wattay airport from which we received samples until 8 May 2022; iii) IPL sampling site; iv) NCLE from which samples have been sent for confirmation or variant analysis (see next paragraph) and v) from other sites that occasionally sent samples to IPL. These other sites are the private French clinic, Center of Infectiology Lao – Christophe Mérieux (CILM), Institute of Diseases Prevention of Lao Army (IDP), Mittapharp hospital and Mahosot hospital (Figure 5).
Figure 5: Origin of COVID-19 samples received at IPL from October 2021 to September 2022.
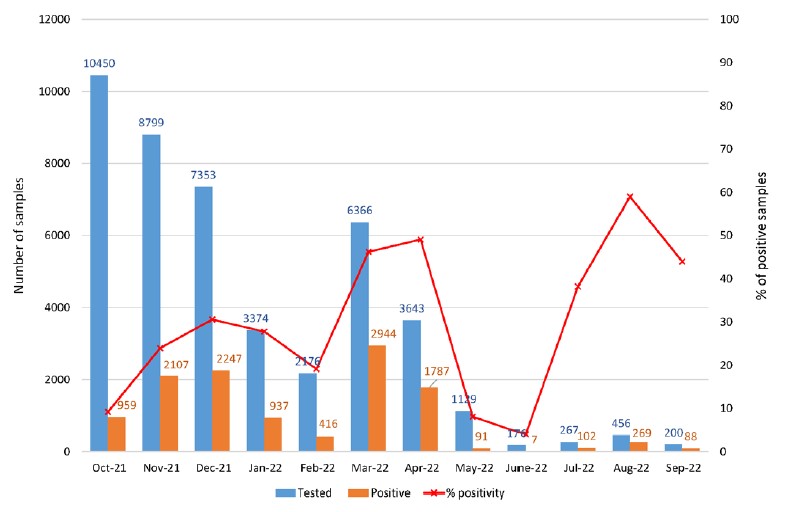
Since June 2022, the number of tested samples decreased significantly reaching less than 500 samples per month, as compared to the end of 2021 when more than 7000 samples were tested every month. However, the percentage positivity between July to September 2022 was still high, probably reflecting a better selection of suspected COVID-19 cases for which swabs have been sent from hospitals to IPL (Figure 6).
Figure 6: Number of tested and positive COVID-19 samples per month.
All RT-PCR results were shared with NCLE and DCDC on a daily basis. NCLE consolidated testing data from all laboratories and reported aggregate data to the MOH. This active surveillance enabled the MOH to track the epidemiologic situation, providing information to track close contacts to limit the spread of the virus. In addition, this information was used for public health decision making.
Surveillance of SARS-CoV-2 variants
In 2020, IPL signed a material transfer agreement (MTA) with NCLE to obtain SARS-CoV-2 RT-PCR positive samples in order to perform surveillance of SARS-CoV-2 Variants of Concern (VOC) and Variants of Interest (VOI). Currently, there are 2 different approaches which are available and recommended by WHO to perform SARS-CoV-2 variants surveillance. The first one is the use of RT-PCR systems that target specific mutations in the spike protein of SARS-CoV-2 that allow differentiation between different VOC and VOI. The second one consists of sequencing either i) full-length genome of SARSCoV- 2; or ii) the full spike gene; or iii) a specific region of spike gene in order to identify the mutation specific to each VOC or VOI.
IPL is currently performing the surveillance of SARSCoV- 2 variants by combining these two approaches on samples selected and sent from NCLE or from the IPL surveillance network. All VOC surveillance results are shared with NCLE, WHO and all partner laboratories immediately, by email and then via a database accessible virtually by various partners. IPL also submits the sequences identified by Sanger sequencing on the GISAID database (https://www.gisaid.org/). Sequencing results allow identification of the circulating variants present in Laos. These results have important implications for the MOH, to support their efforts to track the spread and to reduce the COVID-19 burden. This is in part because different variants present different potential for spreading, and can inform the establishment of different mitigation strategies depending on the situation (opening of quarantine centers, closing schools, restaurants, borders, restrictions of movements between provinces, districts, etc). Thus, the identification of variants was invaluable to the COVID-19 response in Laos.
SARS-CoV-2 sequencing by Sanger technology
From October 2021 to September 2022, IPL attempted to sequence a total of 494 samples, and obtained sequences for 432 samples. For the remaining 62 samples, the laboratory was unable to obtain any amplification product, due to a too low viral load (Ct> 30) or technical issues. Among the 432 obtained sequences, 183 correspond to Delta variant that circulated between June 2021 and March 2022 and was responsible for the biggest COVID-19 wave in Laos. The first Omicron case detected in our samples was in January 2022, and since then 53 sub-variant BA.1, 97 sub-variant BA.2 and 99 sub-variant BA.4 or BA.5 have been identified among the samples submitted for Sanger sequencing (Figure 7). Unfortunately, the targeted region selected for Sanger sequencing corresponding to amino acids 1 to 152 and 313 to 652 of the S protein do not allow the distinction between sub-variants BA.4 and BA.5.
Figure 7: Number of variants identified by Sanger sequencing per month.
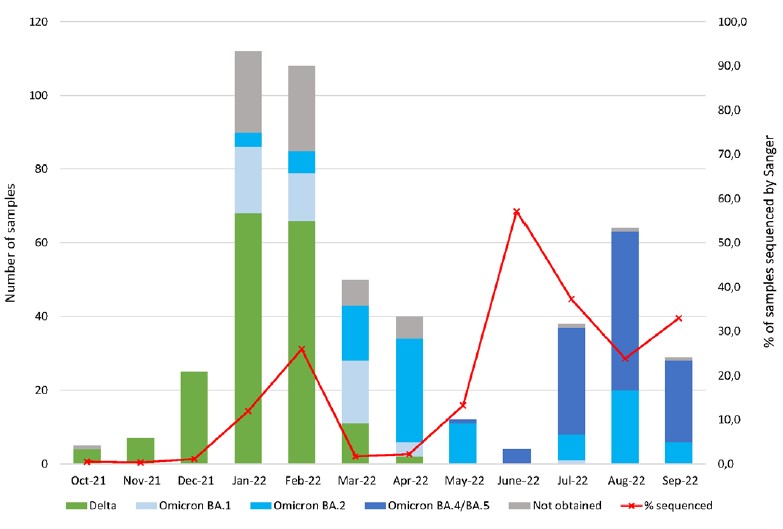
During a few months (October to December 2021 and March and April 2022), due to the high number of samples submitted to IPL for COVID-19 diagnosis, it was very difficult to maintain the Sanger sequencing activity, therefore the percentage of positive samples analysed by Sanger sequencing varies depending on the month (Figure 7).
SARS-CoV-2 variant screening by RT-PCR
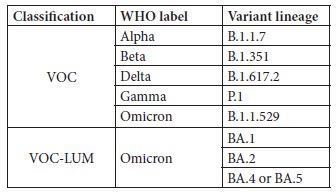
At the beginning of 2022, IPL ordered real-time RT-PCR assay kits intended for the in vitro qualitative detection of SARS-CoV-2 variants in upper respiratory specimens. This assay detects key mutations of VOC, VOI and LUM (lineages under monitoring) classified by WHO. This approach is complementary to the Sanger sequencing to identify VOC. Indeed, with this assay, which combines 2 multiplex RT-PCR (5 targets for each PCR) performed in parallel, it is possible to distinguish the VOC and VOCLUM listed in Table 1.
Table 1: VOC and VOC-LUM detected by the RT-PCR screening assay
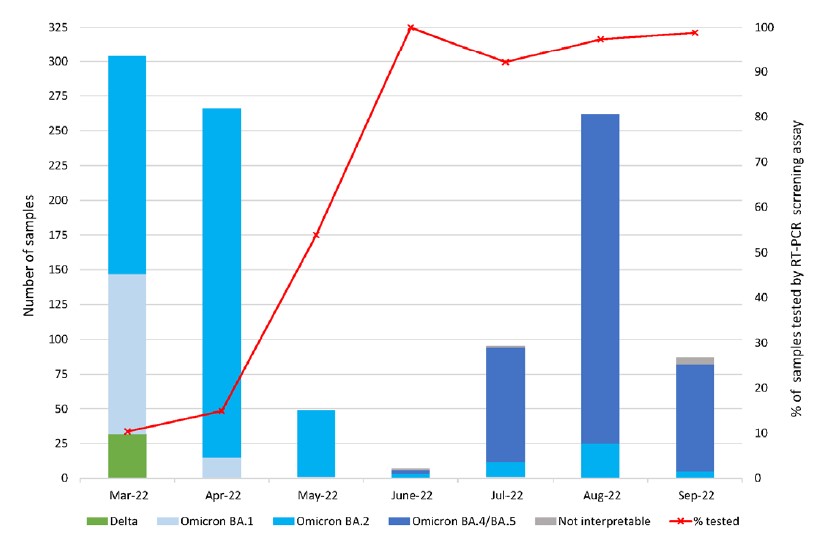
After reception of this new assay, the strategy implemented by the team was to screen the samples with this RT-PCR assay and according to the VOC determination results to select specific samples on which to perform the Sanger sequencing. Thus, between March and September 2022, 1069 samples have been screened by RT-PCR for VOC determination, corresponding to 20.2% of the SARSCoV- 2 positive cases detected at IPL during the same period. In March 2022, we used this assay to screen 10% of SARS-CoV-2 positive samples. The percentage of samples tested by this RT-PCR assay increased gradually during the first months of implementation to reach around 100% from June 2022 onwards (Figure 8).
Among the 1 069 samples screened by this assay, 32 correspond to Delta variant (3%), detected in March 2022, at the end of Delta wave in Laos.
Thus 96.4 % of the samples correspond to Omicron cases with 12.6% of sub-variant BA.1 (n= 132), 46.8% of subvariant BA.2 (n=500) and 37.3% of sub-variant BA.4 or BA.5 (n=399). Less than 1% of the results were not interpretable with this assay (n=7) (Figure 8).
Figure 8: Number of variants identified by RT-PCR screening assay per month.
In March 2022, IPL trained NCLE and CILM teams to use this RT-PCR screening assay and gave each lab reagents in order to be able to perform 200 tests.
Comparison of VOC by Sanger sequencing and RT-PCR screening
The results of VOC characterisation by Sanger sequencing and RT-PCR screening were consistent except for a few cases.
The main advantage of the RT-PCR screening system is the ease of use and rapidness. Indeed, this assay can be performed in 4 hours, compared to 2 days for Sanger sequencing. Moreover, the analysis of the results of RTPCR screening assay takes only a few minutes compared to several hours for Sanger sequencing. However, the RTPCR VOC screening system has some limitations. Firstly, as this system is based on an RT-PCR assay, a mutation in the genome sequence targeted by the primers and/or probes can decrease the efficiency of the RT-PCR and lead to incorrect results. Secondly, when new VOCs emerge they cannot always be detected by the primers specific for the mutations selected by the company who designed the kit. Thus new kits need to be developed by the company, which become available several weeks or months after the emergence of the new VOC.
In contrast, the Sanger approach allows the detection of new mutations and/or variants without any adaptation, so long as the mutation of any new variant is inside the nucleotide region targeted by the PCR. This is why the 2 approaches are complementary and are used together at IPL.
However, as they target either specific mutations for the RT-PCR screening system or a restricted region of the spike gene for the Sanger sequencing, neither can distinguish sub-variant BA.4 and BA.5. This is why, in early 2023, we will implement the full-length genome sequencing by using Nanopore technology. This approach is recommended by WHO for VOC identification and will be a real asset for the country.
Acknowledgments
The Medical Virology and Rabies group would like to thanks Dr. Somphavanh Somlor, Dr Chittaphone Vanhnollat, Phaithong Bounmany, Sitsana Keosenhom, Souksakhone Viengphouthong and Phonesavanh Luangamath for their help in the COVID-19 surveillance activities.







