Origins, natural reservoirs and Interspecies transmission of SARS-CoV-2 and other SARS-like coronaviruses (ONRITS)
 Project Coordinator:
Project Coordinator:
• Dr. Paul Brey, Director, Institut Pasteur du Laos, Vientiane, Lao PDR
• Dr. Khamsing Vongphayloth, Research entomologist, Institut Pasteur du Laos
Staff members:
• Somphavanh Somlor, Research Scientist, Arbovirus & Emerging Viral Disease Laboratory, Institut Pasteur du Laos
• Chittaphone Vanhnollat, Research Scientist, Arbovirus & Emerging Viral Disease Laboratory, Institut Pasteur du Laos
• Khaithong Lakeomany, Technician entomologist, Institut Pasteur du Laos
• Nothasine Phommavanh, Technician entomologist, Institut Pasteur du Laos
• Khammany Oudomsouk, Technician entomologist, Institut Pasteur du Laos
Participating team from the National University of Laos
• Bounsavanh Daongboupha (Chiropteran, Faculty of Environmental Science)
• Vilakhan Xayaphet (Chiropteran Assistant, Faculty of Environmental Science)
• Daosavanh Sanamxay (Rodent, Faculty of Environmental Science)
Outside partners
• National Institute of Hygiene & Epidemiology-Vietnam
• The Pathogen Discovery Laboratory, IP-Paris-France
Funded by the Institut Pasteur du Laos
Period of Project: Extension 2021-2022
Background
Since the discovery of severe acute respiratory syndromeassociated coronavirus (SARS-CoV) in Chinese horseshoe bats (Rhinolophus sinicus) in Hong Kong in 2005, a plethora of molecular epidemiological studies have been carried out in bats and animal species around the world to detect other animal reservoirs of this highly pathogenic CoV (Wong et al., 2019). Interestingly, of the >30 CoVs found in bats, a majority have been found in various species of horseshoe bats (Rhinolophus spp.) from China. A few others have been found in other bat genera such as, Aselliscus, Chaerephon, and Hipposideros.
Given the fact that two SARS CoVhave emerged and cause significant human loss of life in the past seventeen years (SARS and COVID-19), the scientific community postulates that other SARS Coronaviruses are presently circulating in Rhinolophus and other bat populations and could spill-over into human populations in a similar manner to SARS and COVID-19; hence, the surveillance and identification of these novel putative coronaviruses is necessary to mitigate a future pandemic.
A better scientific understanding of the origin, natural history and dispersal of SARS-CoV-2 and SARS-like- CoVs in their natural environment, as well as the mechanisms leading to their interactions and interspecies jumping – Bat-Bat, Bat-Animal and Bat-Human – are paramount to mitigate future spillover events that could result in devastating epidemics like the one we are witnessing today with SARS-CoV-2 in China and around the world.
As mentioned in the IPL Annual report 2021, we set up this study in early 2020 at the beginning of the COVID-19 epidemic. Between 2020 and 2021, bat samples were collected from two provinces of Laos, and results showed that bat-SARS-CoV2-like viruses were detected from samples that were collected in Feung district, Vientiane Province. Herein, we update our activities that were continued between 2021 and 2022.
Objectives
The specific objectives of this project are to; i) assess the presence of SARS-CoV2 and related viruses in Laos and Vietnam wildlife (bats and other mammal communities at the interface with them); ii) search for other SARS-like CoVs using New Generation Sequencing (NGS)-based metagenomic analysis of selected samples; iii) detect past beta-coronavirus infections using specifically developed antibody tests for SARS-CoV2 and closely-related novel viruses.
Methodology
Site selection and time for field sampling in 2022
In August 2022, one field mission of bat collection was made in Feung district, Vientiane province, where the bat-SARS-CoV2-like viruses were found in 2020.
Field collection and bat identification procedure
We follow the same field collection of bats and sampling procedure as detailed in IPL Annual report 2021 except no rodent and other animals were trapped. Laboratory procedure Extraction Total nucleic acids (DNA/RNA) were extracted from bat and rodent anal swabs by using Nuclo Spin®8 Virus (MACHEREY-NAGAL, Germany).
Lab techniques and assay procedure for Pan-corona screening at IPL
cDNA was synthesized using Maxima H minus first strand cDNA synthesis kit (Thermo Scientific) with random hexamers following the manufacturer’s instructions. Pan-corona PCR targeting the RNA-dependent RNA Polymerase gene was used for screening of coronaviruses as previously described by Chu et al. 2011. PCR products of the expected size were directly sequenced on both strands by Sanger sequencing using the nested PCR primers. The sequences obtained were confirmed by similarity analysis using the NCBI BLASTn search (http://www.ncbi.nlm. nih.gov/BLAST).
NGS at IPP
Total nucleic acids (DNA/RNA) for both positive and negative samples and aliquot of anal swabs that were positive following pan-corona PCR at IPL will be sent to IPP for in-depth analysis.
Preliminary results 2022
Bat collection and biological sampling in 2022
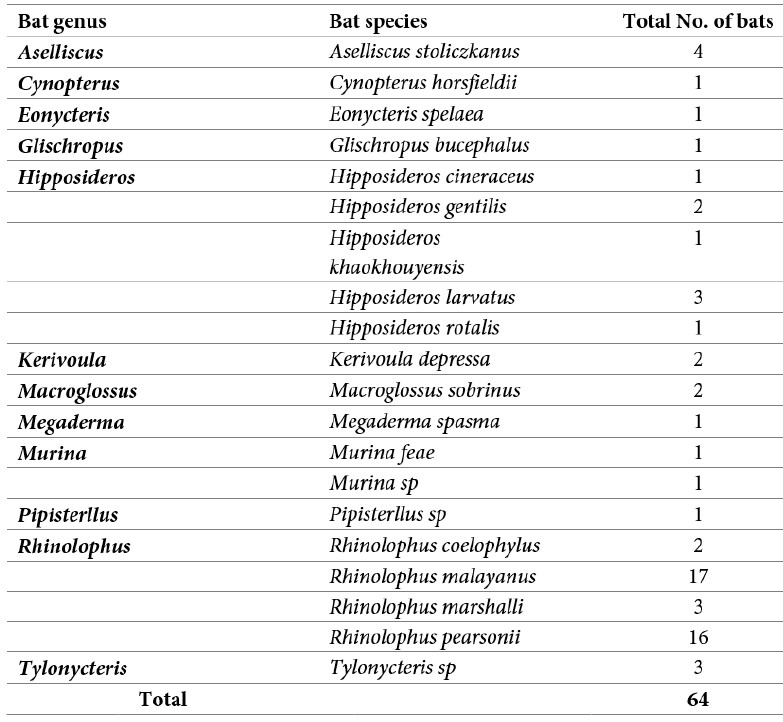
A total of 64 bats were captured from our 4-day collection in Feung district in August 2022 (Table 1). A total of 64 saliva, 60 anal/feces, and 60 urine swab samples were collected. No rodent and other animals were collected in 2022.
Table 1: Total number of bats captured
Pan-corona screening at IPL in 2022
A total of 60 extracted products of bat anal swabs/faeces were screened at IPL by targeting the RNA-dependent RNA polymerase gene. Only one sample was positive from nectar bat Eonycteris spelaea. BLAST analysis of the obtained nucleotide sequence from Sanger sequencing identified as unclassified Betacoronavirus, with 98.52% identity to Bat coronavirus, GenBank accession number MG762606.1, found in the same bat species from China. All samples that are collected during this study will be sent to IPP for continuing further in-depth sequencing.
Summary data of the project between 2020 and 2022
Overall, during the course of this study between 2020 and 2022, a total of 937 bats were examined of which 926 saliva, 863 anal, 322 urine, and 295 blood samples were collected from 6 bat families belonging to 21 genera (Table 2).
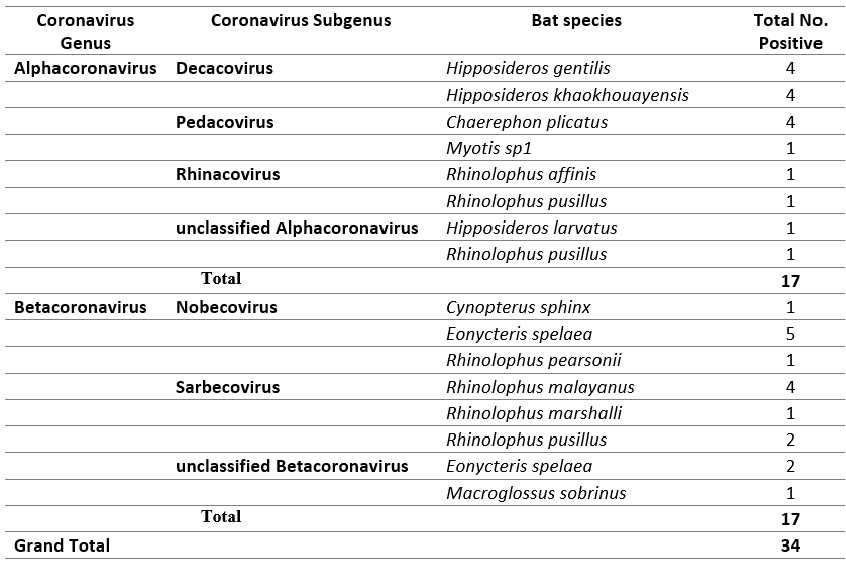
A total of 863 anal swab samples were screened for coronaviruses and 34 were positive. BLAST analysis of obtained nucleotide sequences from Sanger sequencing on the RNA-dependent RNA polymerase gene identified 17 alphacoronaviruses and 17 betacoronaviruses from different bat hosts as show in Table 3 below. So far, a total of 11 Sarbecoviruses were processed for whole genome sequencing at IPP. Five full genomes were obtained and published in 2020 (Temmam S. et al., 2020), while another 6 were not completed. Other remaining alpha and beta coronaviruses will be analyzed using NGS by the IPP team. Isolation from a total of 14 anal swab samples positive for betacoronaviruses was attempted at IPP, but only one (BANAL-20-236) was successful (Temmam S. et al., 2020). As shown in IPL annual report in 2021, the closest bat coronaviruses to human SARS-CoV2 were found from samples collected in Feung district. The results showed that Laotian bat-sarbecovirus RBD was able to bind to human ACE2 through modeling and binding experiments, and Pseudoviruses expressing BANAL-20-236 spike could enter into cells expressing human ACE2. Further experiments such as investigating whether Pseudoviruses expressing BANAL-20-236 spike can directly entry into human cells, and pathogenicity of these viruses are warranted. IPP team will continue to study pathogenicity of BANAL-20-236 using an animal model.
Table 2: Total number of bats captured and biological samples
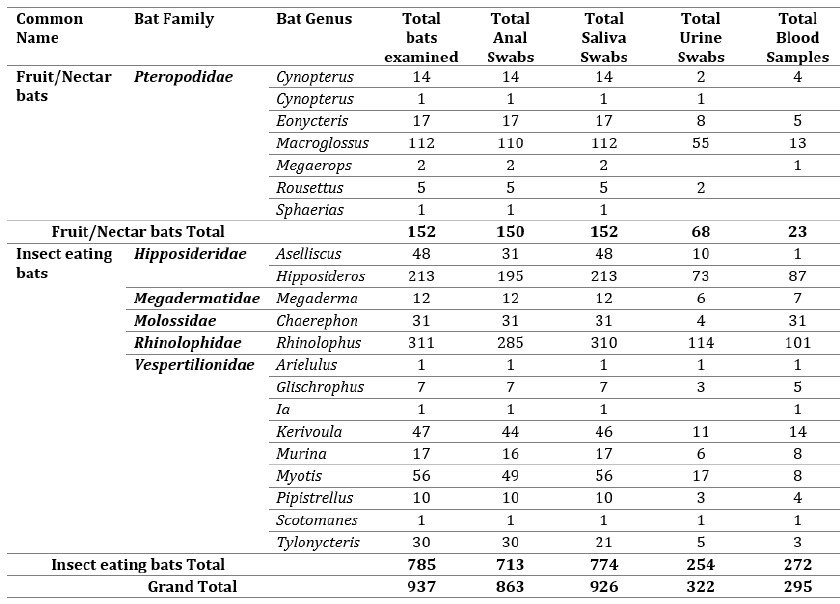
Table 3: Summary results of Pan-corona screening from different hosts from 2020 – 2022
References
– Brunet-Rossinni, A.K. and G.S. Wilkinson. 2009. Methods for age estimation and the study of senescence in bats. Pp: 315-325, in Ecological and behavioural methods for the study of bats, 2nd edition (T.H. Kunz and S. Parsons, eds). The Johns Hopkins University Press, Baltimore, xvii + 901 pp.
– Chu, D. K. W. et al. Avian Coronavirus in Wild Aquatic Birds. Journal of Virology 85, 12815–12820 (2011).
– Chua B.K., et al. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes and Infection, 4:
145–151.
– Corbet, G.B. and J.E. Hill. 1992. The mammals of the Indomalayan region: a systematic review. Natural History Museum Publications and Oxford University Press, 488 pp.
– Csorba, G., P. Ujhelyi, and N. Thomas. 2003. Horseshoe bats of the World (Chiroptera: Rhinolophidae). Alana Books, xxxii + 160 pp.
– Francis, C.M. 1989. A comparison of mist nets and two designs of harp traps for capturing bats. Journal of Mammalogy, 70: 865-870.
– Francis, C.M. 2008. A field guide to the mammals of Thailand and South-east Asia. New Holland Publishers (UK) Ltd and Asia Books Co., Ltd, Bangkok, Thailand, 392 pp.
– Racey, P.A. 2009. Reproductive assessment of bats. Pp: 249-264, in Ecological and behavioural methods for the study of bats, 2nd edition (T.H. Kunz and S. Parsons, eds). The Johns Hopkins University Press, Baltimore, xvii + 901pp.
– Smith,C., C. DeJong and H.E. Field (2010) Sampling small quantities of blood from microbats. Acta Chiropterologica. 12(1): 255-258.Bates, P.J.J. and D.L. Harrison. 1997. Bats of the Indian Subcontinent. Harrison Zoological Museum, Sevenoaks, Kent, England, xvi + 258 pp.
– Wong ACP, Li X, Lau SKP, Woo PCY. Global Epidemiology of Bat Coronaviruses. Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. PMID: 30791586; PMCID: PMC6409556.
Publications
Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D, Xayaphet V, Paphaphanh P, Lacoste V, Somlor S, Lakeomany K, Phommavanh N, Pérot P, Dehan O, Amara F, Donati F, Bigot T, Nilges M, Rey FA, van der Werf S, Brey PT, Eloit M. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022 Apr;604(7905):330-336. doi: 10.1038/s41586-022-04532- 4. Epub 2022 Feb 16. Erratum in: Nature. 2022 Jul;607(7920):E19. PMID: 35172323.
Vongphayloth K, Apanaskevich DA, Lakeomany K, Phommavanh N, Sinh Nam V, Fiorenzano JM, Hertz JC, Sutherland IW, Brey PT, Robbins RG. The Genus Haemaphysalis (Acari: Ixodidae) in Laos: An Update of Species Records and a Review of Taxonomic Challenges. J Med Entomol. 2022 Nov 16;59(6):1986-1992. doi: 10.1093/jme/ tjac075. PMID: 35980598.
Manuscript in preparation
Description of two new species of Phlebotomine sandflies (Diptera, Psychodidae) close to Phlebotomus betisi from Laos with creation of Lewisius subg. nov.







