Arbovirus ecology

Project coordinator: Marc Grandadam
Member of staff: Souksakhone Viengphouthong; Thep aksone Chindavong
The AEVD laboratory was involved in different transversal research programs focused on the identification of hematophagous arthropods species and the evaluation of their potential as vectors of viral diseases. Series of mosquitoes, sandflies ad ticks collected in different provinces and by different approaches were screened by molecular tools to evaluate arthropods infestation rates and prepare biological material for new generation sequencing analysis.
TickMap1.5 – Bio-Lao projects
Research program funded by the US DOD
In collaboration with NMRC-A, Singapore.
Screening of tick specimens (TickMap1.5)
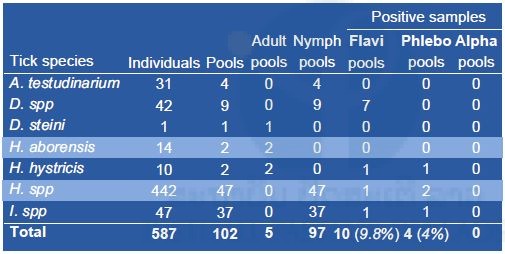
A total of 102 pools, each containing a maximum of 10 specimens, were prepared and submitted to total nucleic acid extraction (Table 3). Field missions allowed the collection of specimens classified in seven species or genera. Lack of reference data on tick species in the Lao PDR complicated the identification. In order to have a better overview of the possible role of ticks in arbovirus transmission, pathogen screening has been extended to the phlebovirus genus (Bunyaviridae). Among the 102 pools of tick specimens screened for arbovirus sequences by means of conventional RT-PCR (Table 3), flavivirus sequences were detected in ten pools (9.8%) distributed across four different species (Table 4). Most of the positives were Dermacentor spp (7/9) followed by Haemaphysalis hystricis (1/2), Ixodes spp (1/37) and Haemaphysalis spp (1/47). Of all samples tested, 4 (4%) displayed a positivity for phleboviruses; and it presented in Haemaphysalis spp (2/47), Ixodes spp (1/37) and Haemaphysalis hystricis (1/2). None of the specimens tested was found positive for alphavirus sequences.
Table 3. Individuals and pool numbers of different tick species collected
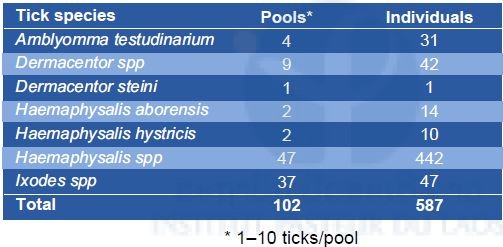
Table 4. Arbovirus screening by RT-PCR targeting flaviviruses, alphaviruses and phleboviruses from tick samples
Flavivirus sequences were evidenced in engorged Dermacentor spp pools (n=4) and Ixodes spp (n=1) pools. Engorged ticks were retrieved from two orders of small mammalians (Rodentia; Scandentia). Flavivirus sequences could also be detected in pools of non-engorged Dermacentor spp (n=3), Haemaphysalis spp (n=1) and Haemaphysalis hystricis (n=1) pools. Phleboviruses were found only in non-engorged tick pools (Table 5). The co-detection of flavivirus and phlebovirus sequences was found within pools of Haemaphysalis spp (n=1) and Haemaphysalis hystricis (n=1).
Table 5. Flavivirus and phlebovirus sequences detected in engorged and non-engorged pools of different tick species

* Collected from Rodentia (3) and Scandentia (2)
Results of RT-PCR screening were used to select candidate samples for virus isolation assays. All tissue supernatant from pools found positive for flavivirus was inoculated on C6/36 and VeroE6, whereas phlebovirus positives were only inoculated on VeroE6 cell lines. Of the ten isolation assays from flavivirus positive samples, three gave a positive result. Of these, only two could be maintained for several passages. Within the four samples found positive for pan-phlebovirus RT-PCR, two led to a positive culture after inoculation on Vero E6 cells. Both could be maintained over three consecutive passages. RT-PCR products for the culture screening were purified to be submitted to Sanger sequencing but this approach failed to provide relevant data for viral species identification. Efforts now focus on amplification of the first passage isolates to verify their stability in culture and obtain sufficient material to attempt a deep-sequencing approach for viral identification and phenotypic characterization.
Screening of arthropods specimen collected in different Lao provinces (BioLao project)
The Medical entomology unit of IPL performed multiple field missions in 2017 to collect new series of ticks, sandflies and mosquitoes in order to improve mapping data and knowledges on the diversity of putative arbovirus vectors. Collection sites in Khammouane, Xayaboury and Bokeo provinces were selected and visited from January to Jun 2017.
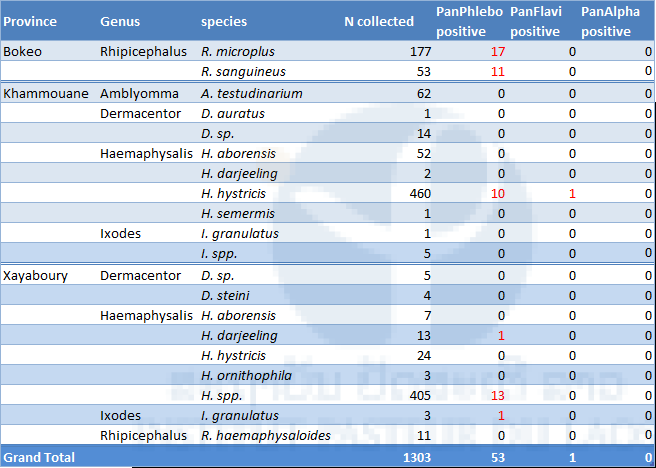
Results of tick collection and pathogen screening are presented in table 6. A total of 1,303 ticks were collected during our field work. Almost all ticks were at the nymphal stage.
The most abundant was Haemaphysalis species (967 specimen), followed by Rhipicephalus spp. (241), Amblyomma spp. (62), Dermacentor spp. (24) and Ixodes spp. (9). Rhipicephalus microplus (17) and R. sanguineus (11) were found positive for phleboviruses in Bokeo province. Haemaphysalis hystricis (10) was also found positive for phleboviruses in Khammouane province. In Xayabouri province, one H. darjeeling, one I. granulatus and thirteen Haemaphysalis spp. were found positive for phleboviruses. Only one specimen (H. histricis) from Khammouane was found positive for flavivirus sequences. None of the tick tested were positive for alphavirus sequences.
Table 6. List of the ticks collected in Laos (2017) in 3 provinces and screened for phelbo, flavi and alphaviruses.
Mosquito collection and pathogen screening
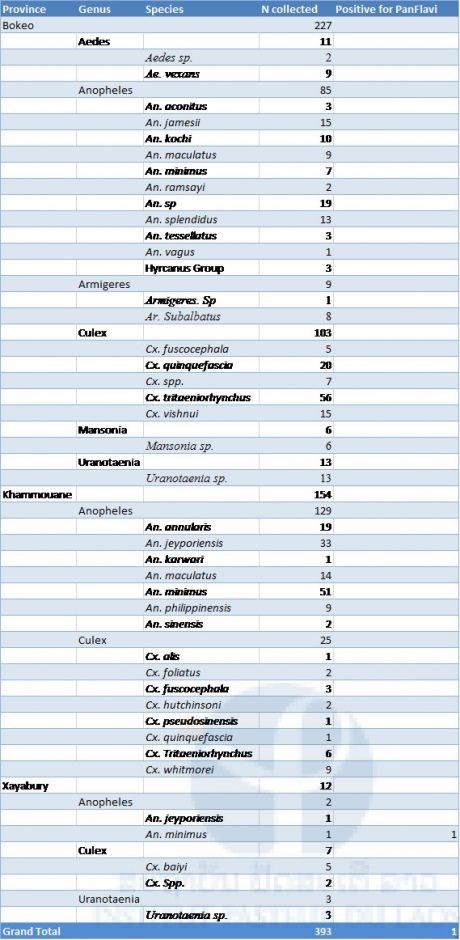
Results of mosquito collection and pathogen screening are presented in the three provinces in 2017 in table 7. A total of 393 mosquitoes were collected during the course of our survey (Table 2). Only females were collected and among them only forty-four were found blood fed. Fifteen different Anopheles species were identified (n=216). The malaria vector An. minimus was the most represented with 51 specimens followed by An. jeyporiensis with 34 specimens. Ten different Culex species were collected (n=135). Culex tritaeniorhynchus and Cx quinquefasciatus were the most represented with sixty-two and twenty-one specimens collected respectively. Culex tritaeniorhynchus is a major vector of Japanese encephalitis in Laos. Only one mosquito from Xayaboury was found positive for alphavirus (An. minimus).
Table 7. List of the mosquitoes collected in Laos (2017) in 3 provinces and screened for flaviviruses.
Sandfly collection and pathogen screening
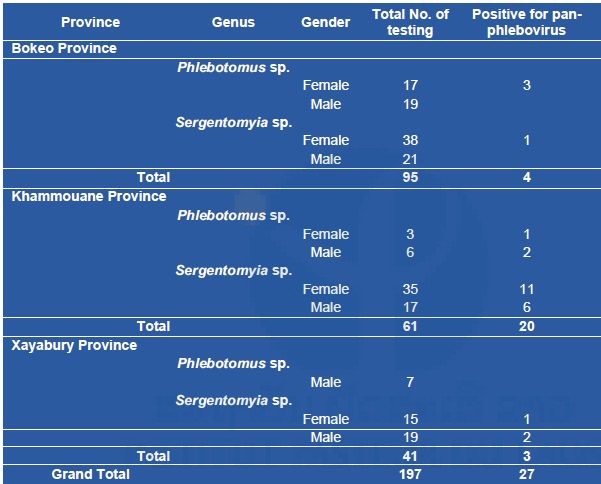
A total of 368 sandflies were captured during our course of collection, of which 197 specimens were cut for identification and screening for pan-phlebovirus. The specimens were identified as Sergentomyia spp. (n=145 of which 88 were females) and Phlebotomus spp. (n=52 of which 20 were females). As show in Table 3, 27/197 specimens were found positive for pan-phlebovirus, 20 from Khammouane province, 4 from Borkeo province, and 3 from Xayaboury province. Both males and females of Sergentomyia spp. and Phlebotomus spp. from Khammouane province were positive (Table 8).
Table 8. List of the sandflies screened for phleboviruses in 3 provinces.
Ticks and sandflies are well known vectors of numbers of human and animal pathogens worldwide. However their distribution and their putative role in human and animal health remain to be determined in numbers of countries including Lao PDR.
Arbovirus screening performed over the Biolao2 project confirmed the high level of positivity of ticks and sandflies for arboviral sequences previously observed by our teams. At this stage, it is not possible to draw a link between the presence of the viral sequences and human and/or animal infections in Lao PDR but some arguments are supporting a potential threat for humans and domestic animal species: (i) the high level and the diversity of ticks and sandflies species and (ii) the detection of positive male specimens suggests a stable maintenance of the virus in sandflies populations through vertical and/or horizontal transmission.
From a virological point of view, it is now of main concern to further document the viral pathogens detected. Specific strategies for preliminary sequencing will be applied to pre-select positive specimens for deep sequencing. In parallel, the remaining biological material will be used for viral isolation assays with the aim to produce viral antigens to develop serological tools that main help screening human populations living in close contact with the putative vectors detected over this project.







