Tick Map 4 project: Mapping of Vectors and Reservoir Hosts in Lao PDR
 Project Coordinator:
Project Coordinator:
Dr. Khamsing Vongphayloth, Research entomologist,
Institut Pasteur du Laos
Dr. Jodi Fiorenzano, Chief of Entomological Sciences,
U.S. Naval Medical Research Center – Asia, Sembawang,
Singapore
Dr. Paul Brey, Director, Institut Pasteur du Laos,
Vientiane, Lao PDR
Staff members:
Khaithong Lakeomany, Technician entomologist,
Institut Pasteur du Laos
Nothasine Phommavanh, Technician entomologist,
Institut Pasteur du Laos
Partners
The Lao-Oxford University-Mahosot Hospital-
Wellcome Trust Research Unit (LOMWRU)
The Pathogen Discovery Laboratory, IP-Paris-France
Funded by the U.S. Naval Medical Research Center-Asia
(NMRC-A) in support of the Department of Defense
Global Emerging Infections Surveillance and Response
System (DoD-GEIS)
Summary
In order to identify common and emerging vectorborne pathogens in Laos, NAMRU-2 Singapore (SG) has established a study to assess the distribution and infection potential of vectors since 2014. Between 2020 and 2021, we continued to conduct tick and other ectoparasite collections in Oudomxay province in the northern part of Laos. The vectors were morphologically identified and pooled. Then nucleic acids were extracted and screened for pan-phlebovirus and pan-flavivirus respectively by conventional nested RT-PCR at IP-Laos; Rickettsia spp. were detected by qPCR at LOMWRU and arboviral pathogens were detected by Next Generation Sequencing at IP-Paris.
A total of 582 ticks and other ectoparasites were collected during the study. No tick samples were positive by panphlebovirus and pan-flavivirus screening. However, results from NGS of two big pools of Dermacentor and Haemaphysalis ticks found 4 viral families known to infect vertebrates including Rhabdoviridae (Yongjia tick virus 2), Reoviridae (Rotavirus), Flaviviridae (Jingmen tick virus), and Unclassified Riboviria (Bole tick virus 4). 121/192 pools were positive for pan-rickettsia for Rickettsia spp. (17kDa by qPCR). However, none of them was positive for R. typhi/Coxiella spp. Our partner, LOMWRU, will continue to try to identify the Rickettsia species. Furthermore, another partner at IP-Paris will continue to perform deep sequencing on the samples collected in the course of this study. From 2021-2022, we will continue our tick study/surveillance in Laos for both ticks and associated pathogens. Future research studies are needed to determine the pathogenicity of these viruses to livestock and humans in Laos.
Background
Vector-borne diseases constitute a significant infectious disease risk for local populations. In Laos, definitive diagnosis is often not available for vector-borne illnesses, so the infectious diseases which are a threat to populations are not well-defined. In order to identify common and emerging vector-borne pathogens in Laos, NAMRU-2 Singapore (SG) has established a study to assess the distribution and infection potential of vectors (including ticks and associated arthropods). In this study, tick and associated arthropod vectors were surveyed from the environment and their associated hosts to provide biological specimens for diagnostic purposes. The samples were transported to the Institut du Pasteur (IPL) laboratory in Vientiane, where a wide range of diagnostic tests can be performed to identify both the vector and pathogens with which they may be infected.
Objectives
Survey and modern identification (ID) of indigenous tick and associated arthropod species
• Collection ID and extraction of vector DNA for submission to and development of the regional repository. This will provide a valuable resource for downstream modern molecular analysis and genotyping.
• Building of local capacities and competencies.
Methodology
Field sites and times
We selected three sites from two villages (Cut and Hauyxang) of Xay district, Oudomxay province (Cut: loc.1: 20.706578°N, 102.113547°E; loc.2: 20.741500°N, 102.107226°E; and Hauyxang: loc.3: 20.763224°N; 102.124142°E) for our surveys. Our first field mission of 9 days was conducted between 11 and 19 February 2021 and the second mission was between 25 July and 4 August 2021.
Field collection procedure
Tick dragging/flagging: Tick dragnets were swept/dragged along the forest ground at approximately 1–2 m intervals before being examined for ticks. Ticks were removed from the sheets using forceps, then transferred to 1.5 ml labeled cryotubes, and stored -20°C. Our dragnet collecting was carried out in all three sites.
Small Mammal Trapping: rodent traps were set for three nights in the Cut village (site 2) and in Hauyxang village (site 3). In both study sites, 50 traps (baited with bananas, sticky rice, or dried fish/meat) were placed in the format of a transect according to the topography. All rodents were released after checking for ectoparasites.
Additional tick collection was carried out by examining domestic animals (cows) in site 2 (Cut village) and in site 3 (Huayxang village). The animal owners were asked to help to examine their animals. Once ticks attached to animals were found, they were collected by direct removal with forceps. All ectoparasite samples were stored at -20°C in the field and transported to Vientiane laboratory (IPL) using dry ice.
Laboratory work
Sample identification and preparation
Ticks were identified under microscopes and grouped in cool conditions (on ice packs) by using reference determination from Dr. Richard G. Robbins of the US Armed Forces Pest Management Board (AFPMB), together with related references from Southeast Asia, Japan, Korea, the Ryukyu Islands (Yamaguti, Tipton et al. 1972), L. E. Robinson keys for genus Amblyomma (Nuttall, Cooper, et al.), and keys from Thailand (Tanskull and Inlao 1989) for adult Haemaphysalis ticks. As there are no morphological identification keys available for pre-imago forms, all larval and nymph stages were grouped into genus. After tick identification and pooling, all information was registered with the Pathogen Asset Control System (PACS) software and all tick samples were stored at −80°C in IP-Laos, Vientiane Capital, for further analysis.
Chigger mites from rodents were mounted on slides using PVA mounting medium. Mite samples were identified using a compound microscope to genus level by referring to the published taxonomic key of Nadchatram & Dohany 1974.
Sample preparation and RNA/DNA extraction
Specimens were placed in 1.5 ml vials containing 1 ml of 1X cold Phosphate Buffered Saline (PBS) and Lysing Matrix A zirconium beads (MP Biomedicals). Tick pools were homogenized for 10 min at a vibration frequency of 25/s in a TissueLyser II system (Qiagen). After grinding, beads and tissues were spun down by centrifugation for 5 min at 3000 rpm. To obtain total nucleic acid (both DNA and RNA) for bacterial and viral detection by polymerase chain reaction (PCR), 100 μl of each pool was extracted and purified by using NucleoSpin® 8 Virus extraction kit following the manufacturer’s protocol. The remaining 400 μl of each pool was stored at –80°C for future pathogen isolation.
Arboviral screening at IPL
Phleboviruses and flaviviruses were screened by conventional nested RT-PCR as previously described (Sanchez-Seco, Rosario et al. 2003; Sanchez-Seco, Rosario et al. 2005).
Bacterial screening
The bacterial screening was carried out in collaboration with LOMWRU, based at Mahosot Hospital, Vientiane. To investigate the occurrence of Rickettsia spp. in ticks, a molecular screening approach targeting the 17kDa gene was taken (Jiang et al. 2004). The presence of Anaplasma spp. and Coxiella was also investigated.
Next generation Sequencing at IP-Paris
A total of 203 pools was sent to IPP for deep sequencing. Our partner will select 26 pools for NGS.
Results
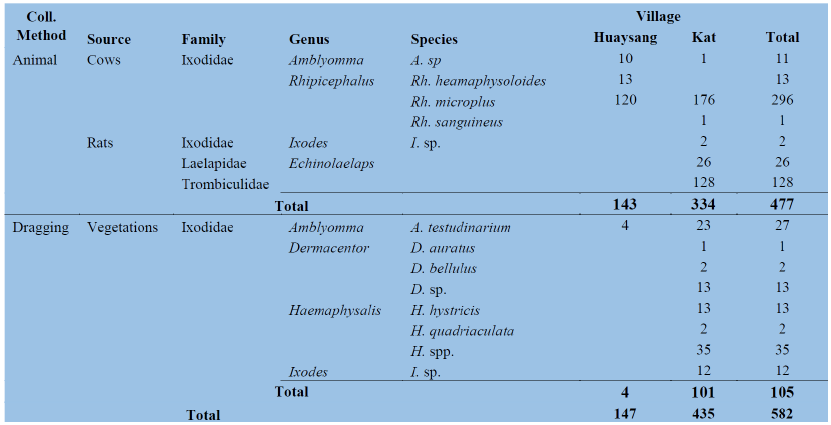
Number, species abundance and composition of ticks and other ectoparasites A total of 582 samples of ticks and other ectoparasites were collected and pooled into 339 pools, of which 477 samples were collected from animals and 105 samples from the dragging method. Five species of ticks were collected from cows and rats whereas 8 tick species were collected from vegetation by dragging. Laelapidae and Trombiculidae were collected only from rats (Table 1 below for more detail).
Table 1: Number of ticks and other ectoparasites collected from different methods by districts.
Preliminary results on the identification of pathogens associated with ticks
Arboviral screening
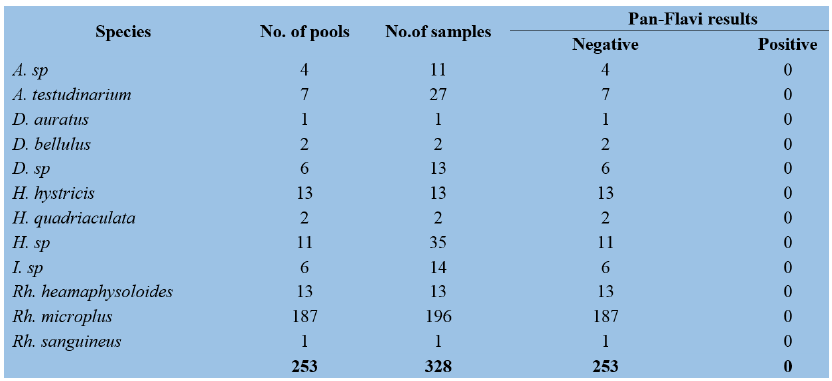
A total of 194 pools (266 tick samples) and 253 pools (328 tick samples) were screened for pan-phlebovirus and pan-flavivirus, respectively by conventional nested RT-PCR. None of them was positive (Table 2 and Table 3). 22
Table 2: Results of Pan-Phlebovirus by RT-PCR
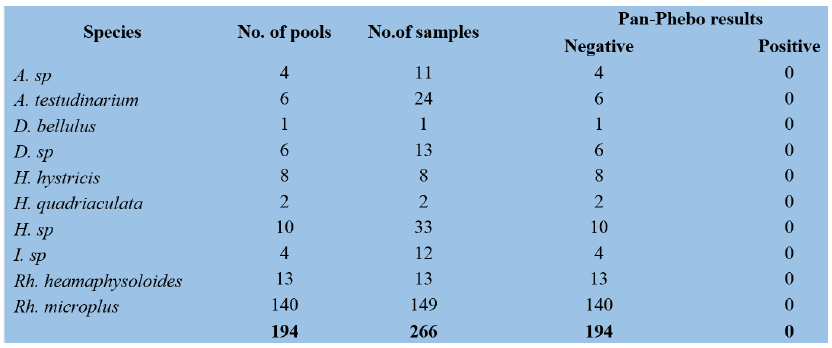
Table 3: Results of Pan-Flavivirus by RT-PCR
Bacterial screening
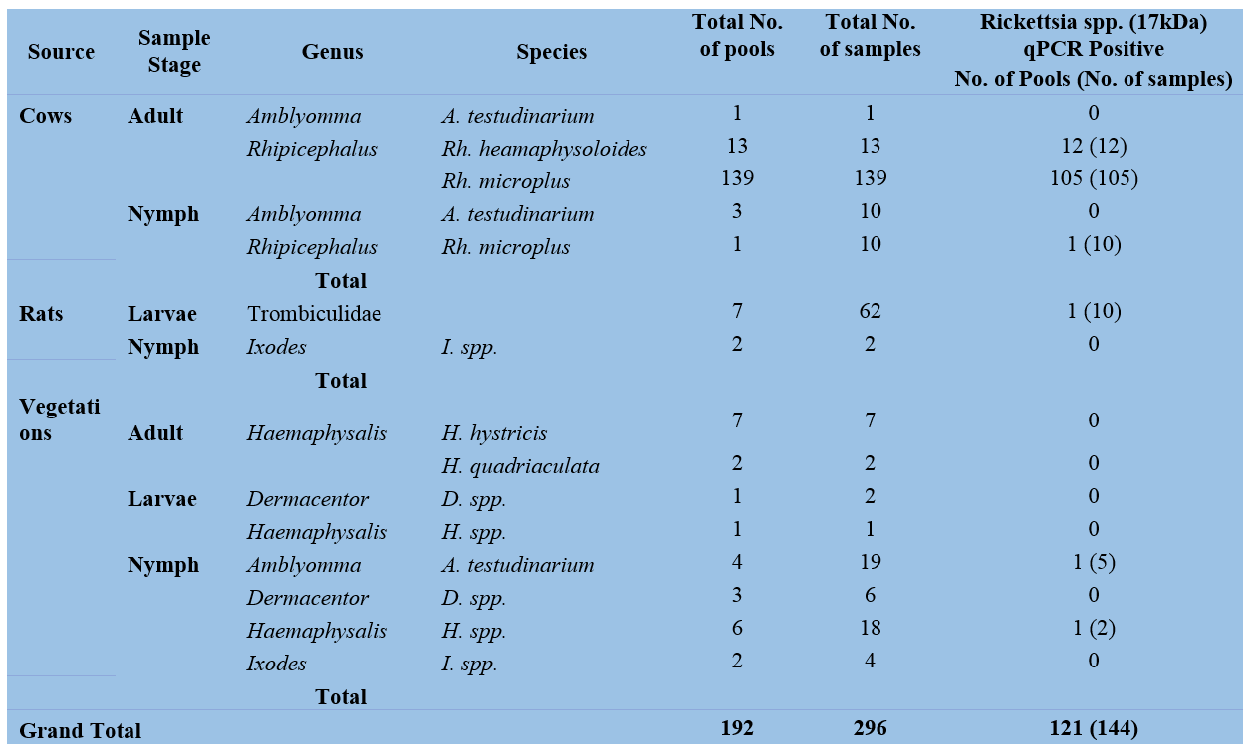
To date, a total of 192 pools containing 296 ectoparasites have been screened for the presence of Rickettsia spp. at LOMWRU. A total of 121 pools were positive for Rickettsia spp. (17kDa) with the highest number of pools positive among Rhipicephalus ticks (Table 4). None of the positive pools (0/100) were positive for R. typhi/ Coxiella spp. The team will continue to try to identify the Rickettsia species.
Table 4: Rickettsial screening from ticks
Next generation Sequencing at IP-Paris To date, deep sequencing has been performed on two pools of Dermacentor (16 ticks) and Haemaphysalis (10 ticks) and other remaining pools have now finished Library preparation as follows:
Library preparation and sequencing by Illumina Nextseq 500
500 2 pools of ticks were selected because of the likeliness of pan phlebo test positivity: Pool 18 (with 16 ticks) and pool 20 (with 10 ticks) (Table 5).
Total RNA extracts were quantified using Qubit RNA High sensitivity assay and quality was checked with an Agilent BioAnalyzer RNA (Table 5 & Figure 1).
Table 5: two pools of ticks selected for the first NGS
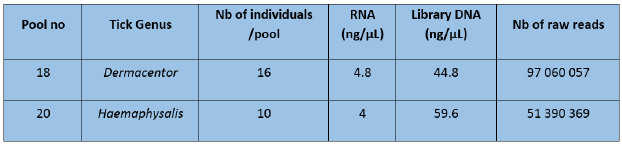
Figure 1: RNA profiles analyzed onto an Agilent BioAnalyzer
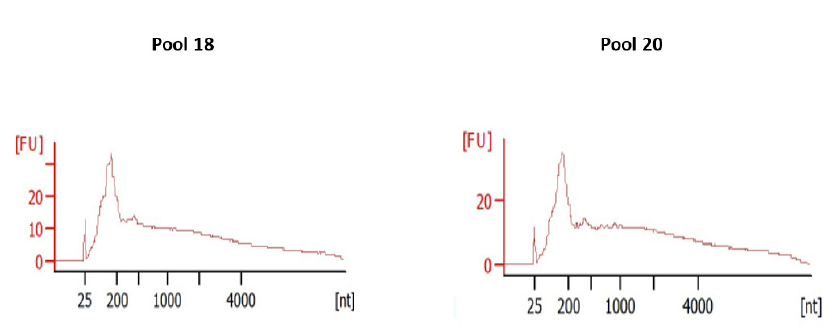
The 2 pools of RNA extracts were used as input for library preparation using the SMARTer Stranded Total RNA-seq kit v3-Pico input mammalian. Quality controls of the libraries comprised a quantification by Qubit DNA High sensitivity assay and characterization of size profiles by BioAnalyzer DNA High Sensitivity chips (Table 5 & Figure 2).
Figure 2: DNA profiles analyzed onto an Agilent BioAnalyzer
Sequencing was then carried out on an Illumina NextSeq 500 sequencer in a single-read 1 x 150 bp.
Bioinformatic Analysis
Total raw reads were processed with an in-house bioinformatics pipeline (Microseek) comprising quality check and trimming reads normalization, de novo assembly using Megahit tool and ORF prediction of contigs and singletons.
A BLAST-based similarity search was performed for all contigs and singletons against the comprehensive and curated protein Reference Viral database (RVDB-prot) followed by a BlastP-based verification of the accuracy of the viral taxonomic assignation against the whole protein NCBI/nr database. A final BLASTN-based verification was performed against NCBI/nt to confirm that no better hit was obtained with non-coding sequences present in NCBI/nt. The quantification of the abundance of each viral taxon was first estimated by summing the length (in amino-acids) of all sequences (contigs and singletons) being associated with this taxon instead of summing the raw number of sequences, to take into account the identification of long viral contigs.
Results from NGS
a. Microbiome data
The Sankey diagrams based on Kraken2 analyses revealed that pool 18 and pool 20 microbiomes are largely represented by bacteria while viruses represent a small minority (Figure 3).
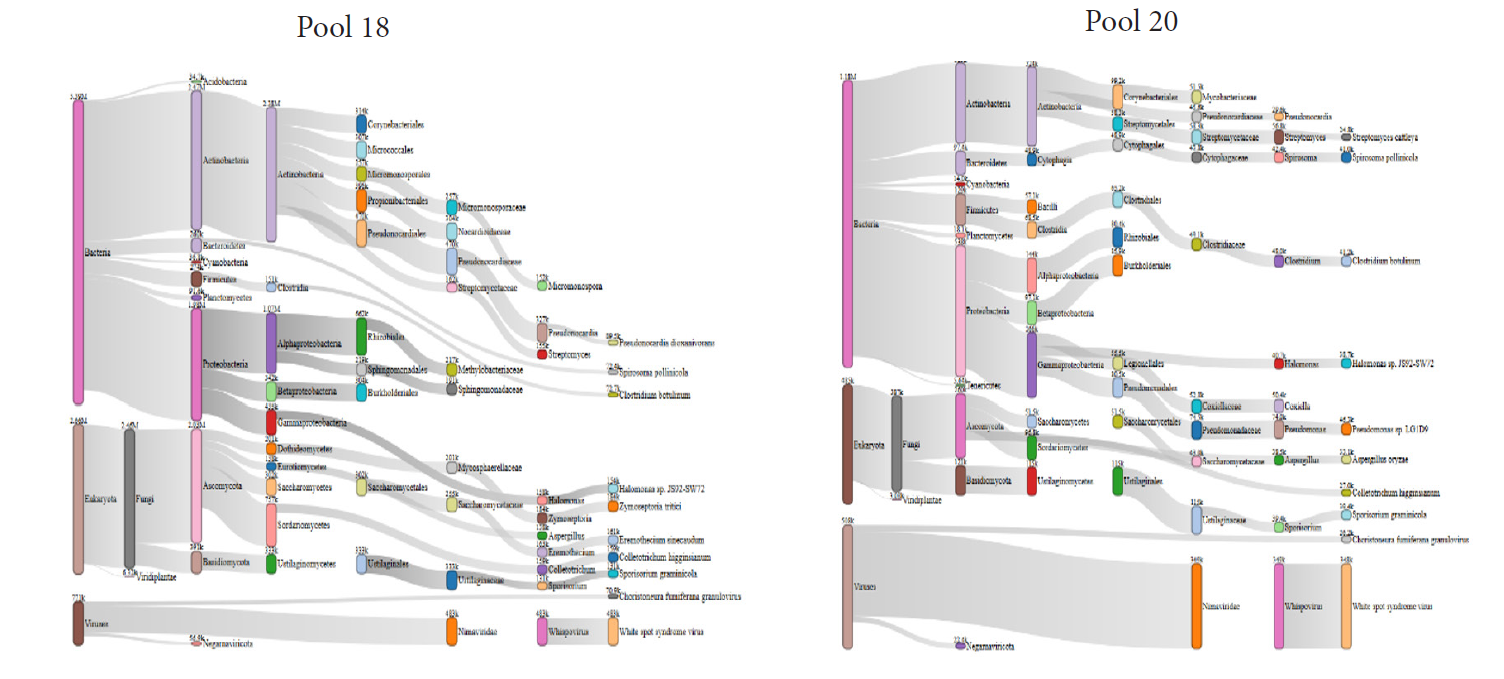
Figure 3: Representation of tick microbiomes using Sankey diagrams based on Kraken2 analyses
b. Virome data
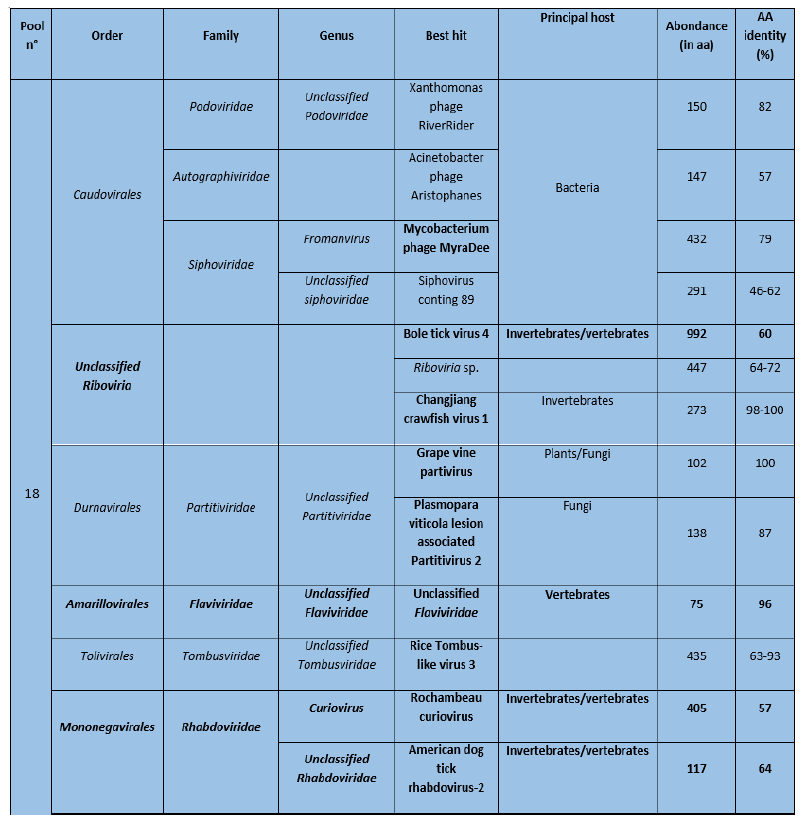
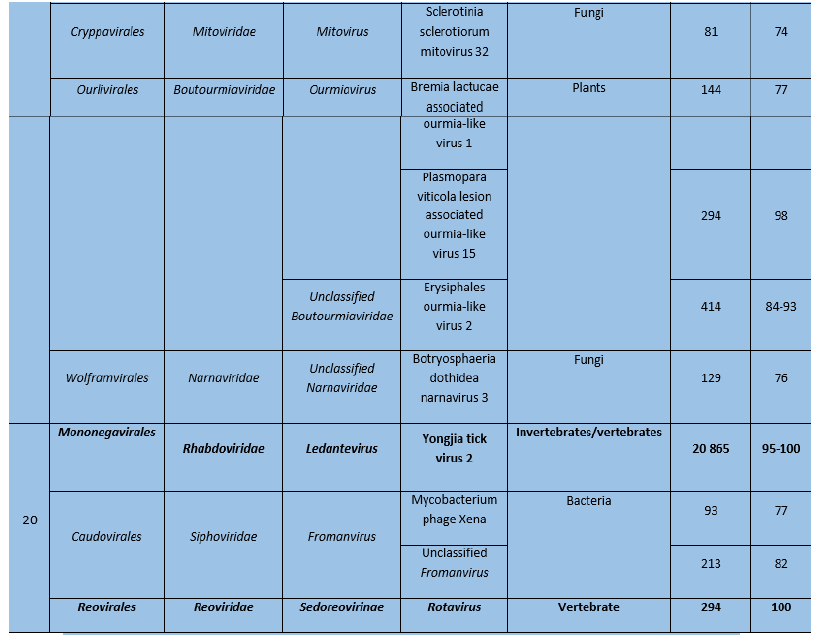
Sequences of many viruses were identified in Pool 18 and most of them usually infect plants and invertebrates (Table 6). In contrast, only sequences of three viruses were identified in pool 20, of which 2 were known to infect vertebrates.
Table 6: Viral sequences identified in ticks using Microseek
c. Mapping of relevant viruses
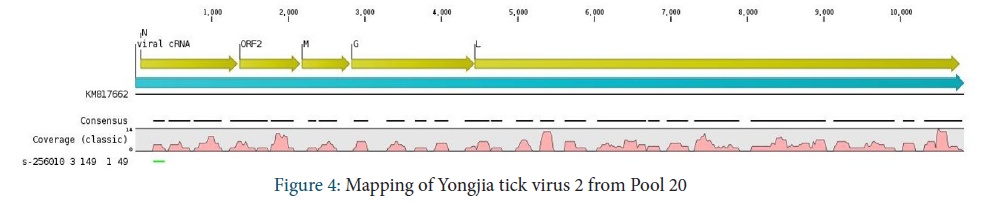
Rhabdoviridae family
Different genera were identified depending on the pool of tick. In pool 20, approximatively 70% of amino-acid sequences that belonged to the Rhabdoviridae were assigned to Yongjia tick virus 2 with 95-100% amino-acid identity. Seventy three percent of genome of the Yongjia tick virus 2 was covered to reference sequence (Figure 4).
In pool 18, 78% of amino-acid belonging to Rhabdoviridae were assigned to Rochambeau virus and 22% were assigned to American dog tick rhabdovirus 2. The latter derive from unclassified rhabdoviruses. It is important to note that these sequences represent a low percentage of amino-acid identity (57% and 64% respectively) and only one read was identified for each best hit. Therefore, we were not able to map these sequences onto the corresponding reference sequences.
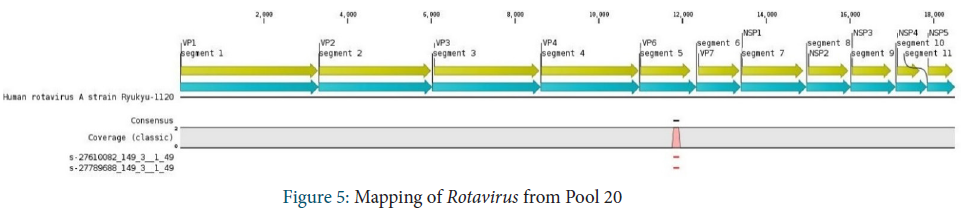
Reoviridae family
Reoviridae family were identified in Pool 20. Only 2 reads were obtained and 100% of these reads were assigned to Rotavirus. They mapped onto a small fragment in the VP6 segment of the virus.
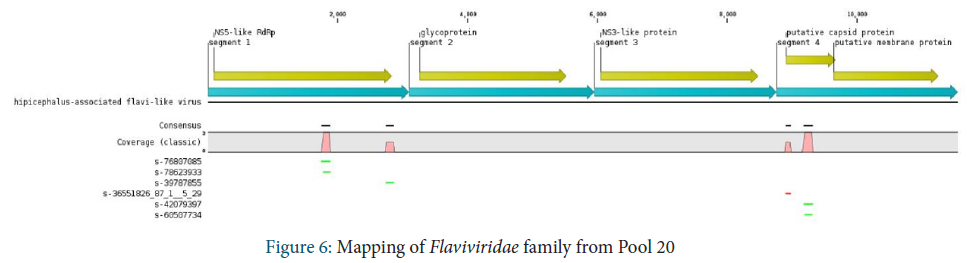
Laviviridae family
The Flaviviridae family is observed in Pool 18. Only a few sequences were identified and assigned to the Jingmen tick virus. These sequences were mapped onto segment 1 coding for the viral polymerase and segment 4 coding for the capsid protein.
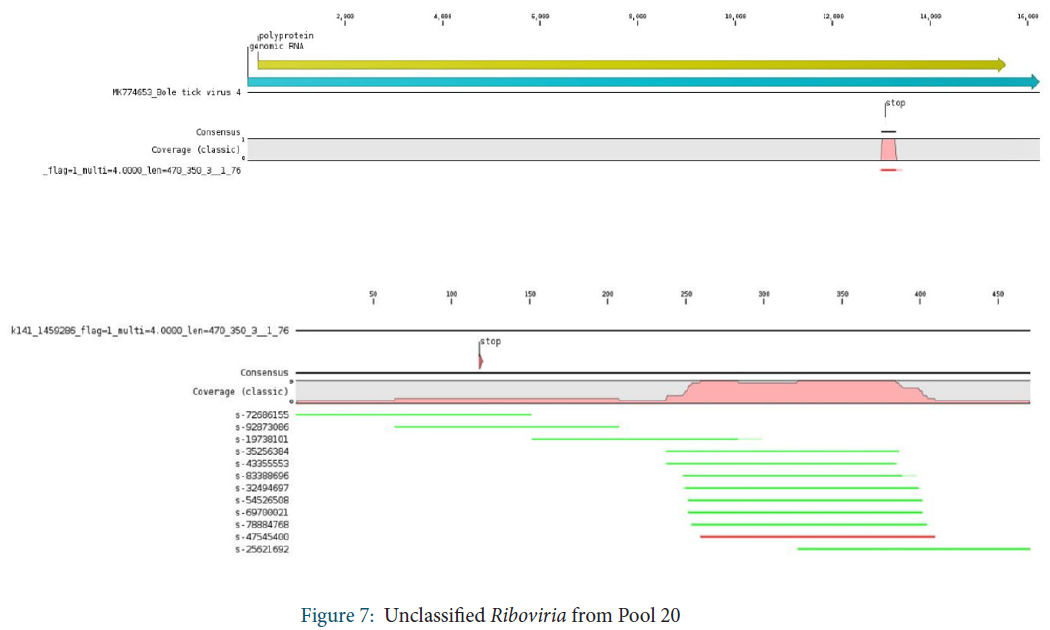
Unclassified Riboviria
Within unclassified Riboviria, sequences assigned to Bole tick virus 4 were identified. A stop codon was observed in the middle of the consensus sequence, but it was covered only by 2 reads.
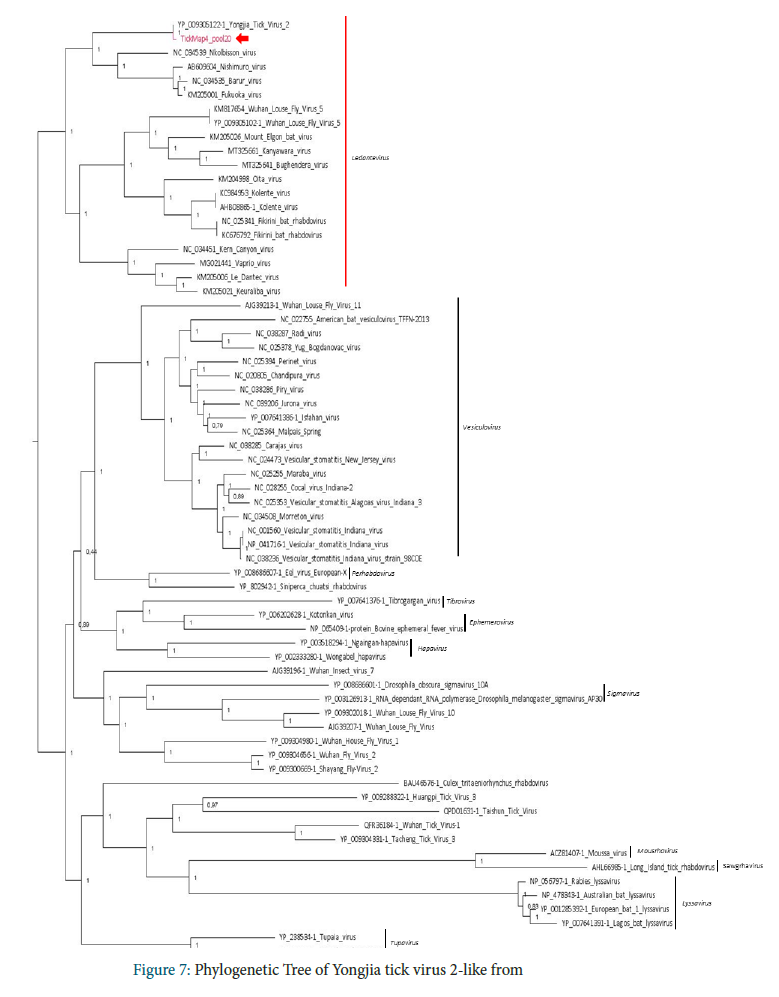
a. Phylogenetic analysis of Yongjia tick virus 2-like from Laos
Phylogenetic analysis performed on the RNA-dependent RNA polymerase amino-acid sequence placed Yongjia tick virus 2-like identified in Pool 20 in the Ledantevirus genus, including Yongjia tick virus 2 previously identified in Haemaphysalis hystricis ticks from China. Interestingly, viruses close to Yongjia tick virus 2 were previously identified in vertebrates: Nkolbisson virus in bats and human cases, Nishimuro virus and Fukuoka virus in ungulates and Barur virus in rodents (Evolution of Genome Size and Complexity in the Rhabdoviridae; Walker P)
Problems Identified during our project and Follow-up Actions
The pandemic of COVID-19 is the only challenge that we have faced so far. However, we have finished all of our field collections on time as planned.
• All samples were stored at between -20°C and -80°C for future pathogen study.
• Our partner, LOMWRU, will continue to identify the Rickettsia species.
• Our partner at IP-Paris will continue to work on deep sequencing
• We will continue the tick study/surveillance in Laos for both ticks and associated pathogens in 2021-2022.
• Future research studies are needed to determine the pathogenicity of these viruses to livestock and humans in Laos.
References
Jiang J., Chan T., Temenek J.J., Dasch G.A., Ching W. and Richards A.L. (2004). Development of a Quantitative Real time Polymerase Chain Reaction Assay Specific for Orentia Tsutsugamushi. American Journal of Tropical Medicine and Hygiene, 70(4): 351-356
Nadchatram, M. and A. L. Dohany. 1974. A pictorial key to the subfamilies, genera and subgenera of Southeast Asian chiggers (Acari, Prostigmata, Trombiculidae). Bulletin from the Institute for Medical Research Federation of Malaysia, 16: 1–67.
Nuttall, G. H. F., W. F. Cooper, C. Warburton, L. E. Robinson, and D. R. Arthur. 1926. Ticks: pt. IV. The genus Amblyomma. Cambridge University Press.
Sanchez-Seco, M.P., et al., Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J Med Virol, 2003. 71(1): p. 140-9.
Tanskull, P. and I. Inlao. 1989. Keys to the adult ticks of Haemaphysalis Koch, 1844, in Thailand with notes on changes in taxonomy (Acari: Ixodoidea: Ixodidae). J Med Entomol 26(6): 573–600.
Yamaguti N., V. J. Tipton, H. L. Keegan, and S. Toshioka (1971). Ticks of Japan, Korea, and the Ryukyu islands. Brigham Young University Science Bulletin, Biological Series 15(1):1.







