Cross-species transmission of poultry pathogens in backyard farms: ducks as carriers of chicken viruses

Project coordinators: Maude Pauly, Claude Muller
Staff members: Latdavone Khenkha, Phonethipsavanh Nouanthong
Background
In Southeast Asia, many risk factors facilitate the cross-species transmission of poultry diseases: large populations of wild and domestic birds, the omnipresence of extensive agriculture, and popular live bird markets. In particular, the village or backyard production system is challenged by enzootic avian diseases. Low hygienic standards, mixing of poultry species and production stages, low-quality feed, uncontrolled bird trade, lack of bird containment and limited access to veterinary and diagnostic laboratory services are drivers for pathogen circulation and cross-species transmission.
Worldwide, the broad range of susceptible hosts and transmission routes represent major challenges to the control of avian viruses. Virtually all bird species are susceptible to Newcastle disease virus (NDV) infection, and wild birds can be a source of infection for domestic poultry and vice versa.
Chickens are the only natural host of chicken anaemia virus (CAV), but other avian and mammalian species may contribute to its spread. Although CAV is mainly transmitted by the fecal-oral route, vertical transmission dramatically impacts the viability of the progeny. Unlike CAV, several bird species are susceptible to infectious bronchitis virus (IBV), an avian coronavirus (CoV). However, other avian CoV strains (e.g. duck CoV) are more species-specific. The emergence of novel recombinants and strains, sometimes in unusual host species, as well as the prevailing lack of pathognomonic signs, further complicates the diagnosis of avian diseases. The clinical course of many avian diseases depends on strain- and host-related factors. Depending on the strain, IBV replicates in the respiratory, urogenital or gastrointestinal tracts of symptomatic or even asymptomatic birds. Inapparent infections are also typical for some pathotypes of NDV, while others lead to high morbidity and mortality.
Vaccination against avian viruses is challenging. Adequate storage and administration of vaccines are difficult in remote settings. Moreover, the protection is short-lived, weak and type-specific, and the vaccines are mostly licensed only for few host species. Due to the high antigenic diversity, the propensity for recombination and the lack of universal cross-protection, a continuous adaptation of vaccine strains to circulating IBV strains is required. These features of avian viruses represent important challenges to backyard farming in rural Southeast Asia.
In Lao People’s Democratic Republic (Lao PDR), poultry rearing is an important source of subsistence, income and high-quality nutritional protein. To assess viral evolution, host range and transmission routes in backyard farms, chickens and ducks from rural areas in Lao PDR were screened for viruses with a broad host range (i.e. NDV and CoV), as well as for a virus with a restricted host range (i.e. CAV).
Results
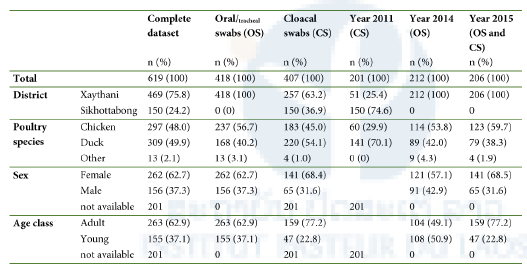
Table 1. Description of sample datasets.
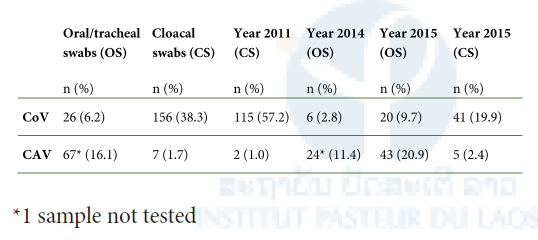
Table 2. Virus detection rates according to sample type and collection year.
Results
CoV detection
Overall, 6.4% (26/404) of the oral/tracheal swabs and 38.7% (156/403) of the cloacal swabs were positive for CoV. High CoV positivity rates were found in cloacal swabs of chickens (27.3%) and ducks (48.2%). Detection rates in the oral/tracheal swabs were significantly lower (9.3% and 2.4%) for both host species. While CoV RNA was consistently found in fecal swabs from every village, viral RNA was only sporadically detected in oral/tracheal swabs. Significantly higher circulation was found in juveniles than in adults (oral/tracheal swabs: 9.7% versus 4.4%, P = 0.04; cloacal swabs: 46.8% versus 12.3%, P < 0.01). IBV-like and duck CoV-like strains were detected in both chickens and ducks. However, IBV-like strains were mostly detected in chickens, and duck CoV-like strains mostly in duck samples (P < 0.001). The most suitable specimen for virus detection depended on the viral strain. IBV-like strains were detected in both swabs of chickens and ducks. In contrast, duck CoVlike strains were detected only in the oral/tracheal swab of a single duck. Only in 2015, both swab types were collected: although overall 9.9% of the oral/tracheal and swabs 20.3% of the cloacal swabs were positive, CoV RNA was detected in both sample types only in 11 cases. Whenever good quality sequences of the partial viral genomes were obtained from both sample types, the nucleotide sequences were 100% identical. The results of the CoV screening are summarized in Table 2.
CAV detection
Overall, 11.6% (47/404) of the oral/tracheal swabs and 1.7% (7/403) of the cloacal swabs were positive for CAV with an apparent local outbreak pattern: CAV positivity was limited to approximately half of the villages (3/6 in 2011 and 4/6 in 2015; no village-related data available for 2014). Positivity was higher in juveniles than in adults (oral/tracheal swabs: 17.5% versus 8%, P < 0.01; for cloacal swabs: 6.4% versus 1.3%, P > 0.05). Although CAV DNA detection predominated in respiratory secretions of chickens (18.1%, 43/237), viral DNA was also detected in oral/tracheal swabs of a few ducks (4/167, 2.4%). Two of the positive ducks were from mixed-species farms located in the same village. While CAV DNA was detected in none of the chicken samples collected in these farms, viral DNA was detected in chickens from other farms in the village. In 2015, CAV DNA was only detected in the two sample types of three birds and the nucleotide sequences obtained from both sample types were 100% identical. The results of CAV screening are summarized in Table 2.
NDV detection
While anti-NDV antibodies were detected in 86.9% (107/123) of chicken sera with no difference in seroprevalence between juvenile and adult birds, virus RNA was detected in none of the swabs.
Viral strain characterization
BLAST and phylogenetic analyses of the partial RdRp gene of CoV, revealed that diverse avian CoV strains co-circulated in rural Lao PDR. A strain clustering with known duck CoV strains was detected in cloacal swabs of 81 ducks and 10 chickens that were collected in 2011 in different villages in both districts. Interestingly, the positive chickens were not from the same farms or villages as the positive ducks. Two different duck CoV strains were detected in cloacal and tracheal swabs of ducks in 2015. Although most Lao IBV strains were obtained from cloacal and tracheal swabs of chickens, some were also detected in duck samples (e.g. AvCoV/ duck/Lao PDR/LAO15_A_11205/2015_MH496841).
All positive ducks were from mixed-species farms or at least from villages with IBV-positive chickens. The vast majority formed a cluster together with the reference strains XDC-2, A2, CK/CH/LLN/111169 and GXNN09032 (accession numbers: KM213963, EU526388, KF411040 and JX897900) from chickens in China. Another Lao strain detected in only one chicken in 2015 belonged to another cluster (AvCoV/duck/Lao PDR/LAO15_A_112551/2015_MH496833) and was most closely related to the reference strain SAIBK from China (accession number: DQ288927). All attempts to sequence the S1 gene of the duck CoV-like strains failed. However, partial S1 gene sequences were successfully obtained from a subset of the IBV-like positive tracheal/ oral and cloacal swabs.
Phylogenetic analyses showed that the Lao IBV-like strains obtained from chickens and ducks may be classified as lineages GI-1, GI-13, GI-19 and GI-25. Most, but not all, of those lineages include strains from China or India. Lao CAV strains clustered in three of the eight recognized CAV lineages, namely lineages 6, 7 and 8. Almost all CAV sequences were obtained from oral/tracheal swabs and, in 2015, most were obtained from samples collected in a single village where strains from lineages 7 and 8 co-circulated. A CAV sequence (MH497005) could only be obtained from one duck sample from the 2014 cohort and it was closely related to the chicken strains. As neither village- nor farm-related information was collected in 2014, it remains unknown whether this duck was raised with CAV-positive chickens. Interestingly, one Lao CAV strain was distantly related to the recognized lineage 1 (i.e. MH496999), but only a relatively small sequence was obtained from this strain (i.e. 578 bp). The vast majority of the Lao strains clustered with sequences from Asia (e.g. China, India, Cambodia and Taiwan).
Discussion
We previously found that 90% of the smallholders in Vientiane Province own poultry with the majority (64%) rearing both chickens and ducks (data not shown). Here, we show high circulation rates of NDV, CoV and CAV, challenging the livelihoods of (semi-)subsistence farmers.
It was difficult to compare our results from Lao PDR to other countries in the region due to the dearth of recent high-quality baseline data on avian diseases in Southeast Asia. It is known that certain avian CoV strains of galliform and non-galliform birds are closely related. The host tropism of CoV depends on the virus strain. In line with previous reports, IBV-like strains were detected in Lao chickens and ducks. Most interestingly, duck CoVlike strains were also detected in both poultry species. Domestic ducks are susceptible to CoV related to viruses of wild waterfowls, which can shed diverse Gammaand Deltacoronaviruses at high rates. Hence, wild and domestic Anseriformes birds may play a role as mixing vessels for diverse CoV strains. In return, chickens probably also represent a minor infection source of duck CoV. Co-circulation of IBV-like and duck CoV-like strains in both species raises concerns for the generation of new CoV variants through recombination.
In contrast to avian CoV, CAV circulation was so far mainly observed in chickens. Interestingly, however, CAV DNA was also detected in a few Lao ducks. It is likely that ducks serve as biological, or at least mechanical, carriers of diverse avian CoV, but also CAV. As most backyard poultry was free-roaming, there are also cross-species interactions with poultry from neighboring farms and with wild birds. This, coupled with the limited sample
size and unknown geographical or commercial origin of the poultry hindered the identification of the infection source.
While the infection status of biological carriers needs confirmation by virus isolation and whole-genome sequencing, mechanical carriers certainly contribute to the viral spread by contaminating the environment. Genetic characterization of numerous Lao CoV and CAV strains from chickens and ducks was attempted. Almost all Lao strains were closely related to strains from Asia, but, as observed previously, the phylogenetic analyses revealed no clear geographic clustering. Most interestingly, the partial genomes from virus strains detected in ducks and chickens were closely related, which further highlights the high risk of cross-species transmission in this setting.
The current nomenclature of IBV genotypes is based on the complete S1 unit of the spike gene characterized by a high frequency of random mutations. This, as well as low viral loads in the samples, explains why the molecular characterization of the S1 unit of all Lao duck CoV and many IBV failed. In addition, recombination between IBV and duck CoV, favored by their cocirculation, may also be responsible for sequencing failure. By referring to IBV-like and duck CoV-like strains, we highlight the uncertainty that remains with regard to the strain classifications as viral genomes were only partially sequenced. In the future, unbiased full genome sequencing is needed to confirm not only strain classification but also to identify possible recombination events.
CoV and CAV co-circulated in approximately 30% of the villages, and nucleic acids of both viruses were detected in four young chickens. Even if none of the Lao poultry was obviously symptomatic, co-circulation of both viruses often increases disease severity. While persistent infections with asymptomatic shedding are typical for young poultry, older birds tend to be resistant to clinical disease, but remain susceptible to infection. Silent shedding complicates diagnosis and facilitates prolonged co-circulation of different viral strains. The latter represents a recognized risk factor for the emergence of novel strains and of recombinants adapted to new host species.
The impact of the circulating CAV or IBV strains on duck production remains unknown. While most reports suggest low pathogenicity of IBV for ducks, a novel IBV strain with clear pathogenicity for ducks was detected in China, also highlighting the need for new IBV vaccines and continuous surveillance. Although detection rates of CAV were highest in tracheal swabs, CAV detection was successful usually only in one of the two swab types, likely due to differences in infection stages and/or viral strain. Thus, both swab types should be tested for CAV to diagnose an infection or assess virus prevalence with high confidence in all host species, including ducks. A similar approach is already recommended for IBV surveillance, but may not be necessary for duck CoV. In fact, this avian CoV was mainly detected in fecal samples.
Cross-species transmission of NDV could not be evaluated in this study. None of the swab samples were positive by multiplex real-time RT–PCR targeting both NDV class I and class II strains. Since CoV, another RNA virus, was successfully detected in the same samples, it is unlikely that sample transport and storage conditions affected NDV detection adversely. However, serological evidence of NDV circulation among Lao chickens was obtained. As the ELISA is only suitable for screening chickens, applying an ELISA with a broader host range would be important to assess the role of ducks in NDV epidemiology in Lao PDR.
Conclusion
Our study shows that backyard farming of mixed poultry species facilitates cross-species transmission and represents an ideal breeding ground for new virus strains. To reduce the risk of infection in backyard settings, improved biosecurity, bird containment, separation of poultry species and sick birds, tailored vaccination programs and continuous surveillance are warranted. Improving the control of avian viruses will not only benefit the birds but will also contribute to the economic sustainability of smallholders. This study was published in the journal Avian Pathology and communicated with our partners at the Faculty of Agriculture, National University of Laos.
Phylogenetic analysis of the partial RNA-dependent RNA polymerase gene of avian Gammacoronaviruses Bayesian analyses of a 329 nt long alignment comprising unique, partial RNA-dependent RNA polymerase (RdRp) gene sequences of 208 coronaviruses (CoV) strains. All available RdRp sequences from reference strains of Infectious bronchitis virus (IBV) were included (in bold and italic) as well as a selection of unique GenBank sequences. The phylogenetic relationship of every unique Lao sequence (in red) to GenBank sequences is shown. When identical sequences were retrieved from several samples, this is shown by indicating the frequency of detection behind the strain name (e.g. “*8”). The sample material from which the sequences were obtained is also displayed (CS: cloacal swab and OC: oral/tracheal swab). Only the posterior probability (pp) values of well-supported nodes (pp>0.7) are shown. The sequences were named, if the information was available, according to the following nomenclature: AvCoV/host/country/specimen id/year_ Genbank accession number. For the IBV reference stains also the lineage as determined before based on complete spike sequences is displayed (e.g. “GVI-9”).
Bayesian analyses of a 977 nt long alignment comprising unique, partial VP1 gene sequences of 129 chicken anemia virus (CAV) strains. The selection of GenBank sequences included in the analysis and also the CAV classification are based on a recent publication. The phylogenetic relationship of every unique Lao sequence (in red) to GenBank sequences is shown. Only the posterior probability (pp) values of well-supported nodes (pp>0.7) are shown. The sequences were named, if the information was available, according to the following nomenclature: Genbank accession number|country|year.








