Tick vector mapping and pathogen characterization study in Laos 2 (Tick Map 2 project)
Funded by the U.S. Naval Medical Research Center-Asia (NMRC-A) in support of the Department of Defense Global Emerging Infections Surveillance and Response System (DoD-GEIS)

Project coordinator:
+ Dr. Paul Brey, Director, Institut Pasteur du Laos, Vientiane, Lao PDR
+ Dr. Ian Sutherland, Chief of Entomological Sciences, U.S. Naval Medical
+ Dr. Jeffrey Hertz, Chief of Entomological Sciences, U.S. Naval Medical Research Center – Asia, Sembawang, Singapore
Member of staff:
+ Khamsing Vongphayloth, Research entomologist, Institut Pasteur du Laos
+ Khaithong Lakeomany, Technician entomologist, Institut Pasteur du Laos
+ Nothasine Phommavanh, Technician entomologist, Institut Pasteur du Lao PDR
Collaborators:
+ Marc Eloit , Virologist, Pathogen Discovery Unit, Institut Pasteur Paris
+ Paul Newton and Matthew Robinson, LOWMRU Wellcome Trust Vientiane Lao PDR
+ Marc Grandadam, Arbovirology Lab, Institut Pasteur du Laos
Background
Vector-borne diseases constitute a significant infectious disease risk for for local populations. In Laos, definitive diagnosis is often not available for vector-borne illnesses, so the infectious diseases, which are a threat to rural and urban populations are not well defined. In order to identify common and emerging vector-borne pathogens in Laos, NMRC-A Singapore (SG) has established a study to assess the distribution and infection potential of ticks and tick-borne pathogens. In this study, tick vectors are surveyed to provide biological specimens for pathogen screening. The samples are transported to the Institut du Pasteur (IPL) laboratory in Vientiane, where a wide range of tests can be performed to identify both the arthropod species and the pathogens with which they may be infected. In order to understand the infectious disease threats in a range of environments in Laos, IPL must collect and screen specimens from several sites and provinces throughout Laos. Further characterization and targeted analyses of positive pathogen samples we have already collected from our first tick study has also been carried out. This study is known as the “Tick vector mapping and pathogen characterization study in Laos” (TickMap 2).
Nakai District, Khammouan Province, is uniquely positioned to serve as a study site for TickMap 2 and includes a large protected area bordering Vietnam in south-central Laos, a region in which many causes of vector-borne illness are not diagnosed, and from which new infectious diseases may emerge. All local staff is already familiar with the requirements of study specimen identification, sample collection, and the principles of vector biology research.
Based on the data and unprecedented success of our prior and ongoing initial phase project lines, including the recording of at least four new species of tick (Vongphayloth et al. 2016), the potential discovery of three new species of rickettsia (Taylor et al. 2016), and a new tick–virus interaction that has never previously been described to science, we are initiating the TickMap 2 project to complement and augment our prior and ongoing tick research efforts in Laos.
In this report, we update all our data on field collection missions, arboviral–bacterial screening, and molecular identification of ticks from the study.
Objectives
+ Survey and modern ID of indigenous tick species distribution.
+ Collection, morphologic identification, extraction of vector DNA for submission, and development of regional repository. This will provide a valuable resource for downstream modern molecular analysis and genotyping.
+ Building of local capacities and competencies.
Methods
Field site collection
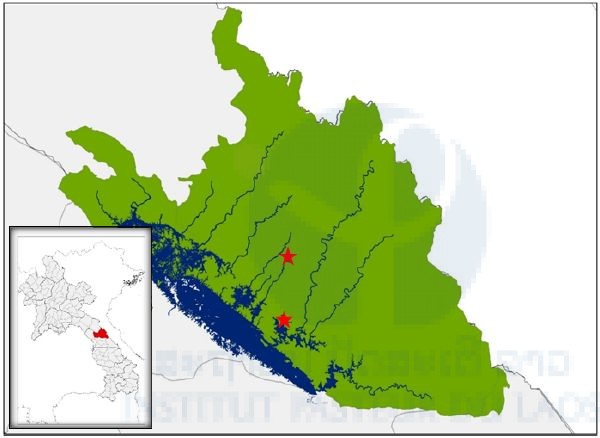
Our first field collection of ticks was carried out between the 12th and 19th January, 2017 in the Nakai Nam Theun National Protected Area (NNT NPA), known as the Watershed Management and Protection Authority area (WMPA), located in Nakai District, Khammouan Province, Laos. Ticks were sampled in two main sites: the Korbong village area (location: 17.879643°N, 105.389993°E), and the Namnoy River area (location: 17.768548°N, 105.381989°E). In the Korbong village area, ticks were collected in secondary forest close to the rice fields of the villagers whereas in the Namnoy River area, ticks were collected inside the primary forest area. The second field collection of ticks was carried out between the 20th and 25th of February 2017 in the Namnoy River area only.
Tick sampling procedure
A dragging method was used for tick sampling from forest floors and vegetation. These standard dragnets were swept along the forest ground at approximately 1–10 m intervals around natural animal trail ways before being examined for ticks. Ticks were removed from the sheets using forceps and transferred to 1.5 ml labeled cryotubes (using PACS labeling and specimen tracking system Black and Vetch).
All live tick samples were then transported to the Nakai field laboratory. In the lab, ticks were placed in the freezer at −20°C for 10 minutes. Then adults, nymphs, and tick larvae were separated, counted, and subsequently transported to IP-Laos in Vientiane Capital and stored at −80°C until processing for further analysis (species identification and pathogen detection). All study sites were data-logged for full GPS parameters.
Tick identification
Ticks were identified and grouped under microscopes in cooling conditions (on ice packs) by using reference determination from Dr. Richard G. Robbins of the US Armed Forces Pest Management Board (AFPMB), together with related references from Southeast Asia, Japan, Korea, the Ryukyu Islands (Yamaguti, Tipton et al. 1972), L. E. Robinson keys for genus Amblyomma (Nuttall, Cooper et al.), and keys from Thailand (Tanskull and Inlao 1989) for adult Haemaphysalis ticks. As there are no morphological identification keys available for pre-imago forms of Haemaphysalis spp., larval and nymph stages in this genus were grouped by using their main morphological characteristics, especially the capitulum. After tick identification and pooling, all information was registered with the Pathogen Asset Control System (PACS) software and all tick samples were stored at −80°C in IP-Laos, Vientiane Capital, for further analysis.
Laboratory procedure for pathogen screening and molecular tick identification
Sample preparation and RNA/DNA extraction
Tick samples were pooled into groups of one to ten by species or genus, sex, stage of development, collection period, and site. Specimens were placed in a 1.5 ml vial containing 1 ml of 1X cold Phosphate Buffered Saline (PBS) and Lysing Matrix A zirconium beads (MP Biomedicals). Tick pools were homogenized for 10 min at a vibration frequency of 25/s in a TissueLyser II system (Qiagen). After grinding, beads and tissues were spun down by centrifugation for 5 min at 3000 rpm.
To obtain total nucleic acid (both DNA and RNA) for bacterial and viral detection by polymerase chain reaction (PCR), 100 μl of each pool was extracted and purified by using NucleoSpin® 8 Virus extraction kit following manufacturer’s protocol. The remaining 400 μl of each pool was stored at –80°C for pathogen isolation.
Arboviral viral screening
Multiple sets of primers have been selected and tested for screening arthropod specimens for the presence of phlebovirus, flavivirus, and alphavirus sequences by means of conventional nested RT-PCR as previously described (Sanchez-Seco, Rosario et al. 2001; Sanchez-Seco, Rosario et al. 2005).
Next generation sequencing on tick samples
RT-PCR screening targeting three main arbovirus genera evidenced the presence of genome fragments in some tick samples. Until now, direct sequencing strategies have not allowed fort preliminary identification of the sequence detected as a probable consequence of the use of degenerated primers for both RT-PCR and direct amplicon sequencing. A collaboration with Dr. Marc Eloit (Pathogen discovery and NGS platform at the Institut Pasteur in Paris) has been initiated to solve the sequencing priming problem and increase the chances of identifying new arboviruses by accessing larger or complete genome sequences. This platform was selected as the staff had good experience in the preparation and virome analysis of ticks.
Bacterial screening
To investigate the occurrence of Rickettsia spp. in ticks in Laos, a molecular screening approach targeting the 17kDa gene was taken (Jiang et al. 2004). The presence of Leptospira spp., Anaplasma phagocytophilum, Ehrlichia chaffeensis, Coxiella burnetii and other Ehrlichia spp., Neorickettsia spp., and Anaplasma spp. was also investigated. The bacterial screening was carried out in collaboration with the Lao-Oxford University-Mahosot Hospital-Wellcome Trust Research Unit (LOMWRU), based at Mahosot Hospital, Vientiane.
Statistical analysis
Infection rate (IR) was calculated using Excel add-in software for mosquito surveillance developed by the CDC. Estimates of the IR are usually presented as the number of infected mosquitoes per 1,000 tested. The simplest estimate, the minimum infection rate (MIR), is calculated: ([number of positive pools / total specimens tested] x 1000), with the data representing a single species or species group collected over a time period and geographic area relevant to the goals of the surveillance program (https://www.cdc.gov/westnile/resourcepages/mosqSurvSoft.html).
Molecular tick identification using the COI – barcode region of the mtDNA
For accurate molecular identification of ticks, we tested the primers LCO/HCO (Folmer et al. 1994) and FF1/R1 (Polsomboon et al.) to obtain a fragment of cytochrome oxidase subunit I (COI), barcode region. Each 15 µl of PCR solution contained 7.5 µl of 2X PCR Master Mix (Promega), 0.6 µl of each primer (10 µM), 1.5 µl of DNA, and water. We followed three different thermocycling programs as previous described (Foster et al. 2013, Rothen et al. 2016, Wang et al. 2012). To confirm the amplification through PCR, we ran the agarose gel 1.5% to analyze our amplification products.
Results
Number of ticks collected and species composition
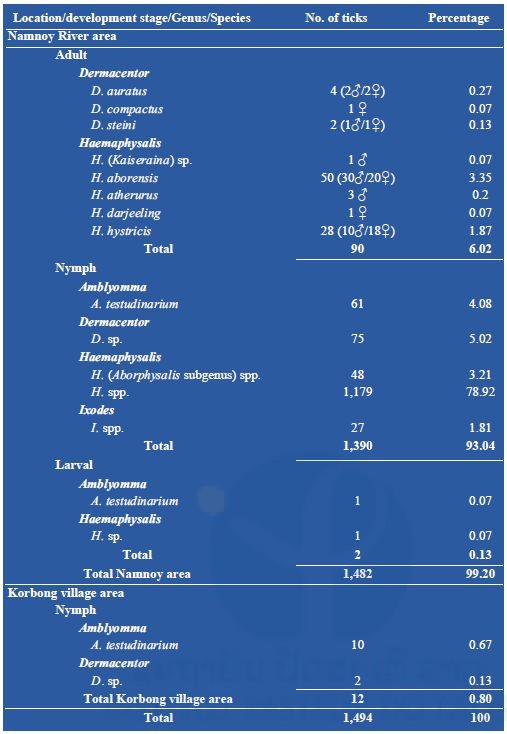
A total of 1,494 ticks were collected during our fieldwork missions. Among them, 1,482 (99%) were collected from the Namnoy area; only 12 (0.8%) were collected from the Korbong area where the collection site was secondary forest. A total of 90 (6%) adults were collected by the dragging method, all from primary forest in the Namnoy area and were identified as Dermacentor auratus (4), D. compactus (1), and D. steini (2); Haemaphysalis aborensis (50); H. atherurus (3), H. darjeeling (1), H. hystricis (28), H. (Kaiseraina subgenus) sp. (1). Nymphs represented the main part (1,402/1,494 or 93.84%) of the total specimens collected belonging to four genera: A. testudinarium (71), Dermacentor spp. (77), Haemaphysalis spp. (1,227) and Ixodes spp. (27). Only two larvae were collected (see Table 1 below for the detail). For the nymph identification, Amblyomma nymphs were classified as A. testudinarium (61), Dermacentor nymphs as D. spp. (75), Haemaphysalis nymphs as H. spp. in the Aborphysalis subgenus (48) and H. spp. (1,179), and Ixodes nymph as I. spp. (27).
Table 1: Total number of ticks collected
Arboviral screening
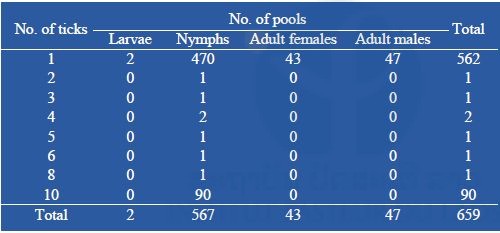
From two field collections, a total of 1,494 ticks were collected and pooled into 659 pools containing 1–10 tick specimens according to collection date, species, and stage of development (n = 659; 567 pools of 1–10 nymphs, 43 pools of 1 adult female, and 47 pools of 1 adult male (Table 2). Arboviral screening was performed by submitting tick nucleic acids (n=659 pools) to pan-flavivirus, pan-alphavirus, and pan-phlebovirus RT-nested PCR.
Table 2: Number of pooled tick collected during our surveys submitted to arboviral screening
Of the total specimens collected (659 pools), 33/659 (5%) pools (78 ticks) were positive for pan-phlebovirus, 5/659 (0.7%) pools (5 ticks) were positive for pan-flavivirus and only 1/659 (0.1%) pool (1 Haemaphysalis nymph) was positive for pan-alphavirus. All positive tick samples were collected from the Namnoy area. None of the samples from Korbong area were positive for any arboviral screening.
As show in Table 3 below, the most positive arbovirus was pan-phlebovirus (33/659 pools) with the highest infection rate (IR) (estimated per 1000 tick samples tested) of 22.40 (95% Confidence Interval [CI]: 15.87-30.74) for the total number of ticks tested. Among pan-phlebovirus positives when analyzed by genus group, Amblyomma nymphs had the highest IR of 309.45 (95%CI: 216.12-415.42) followed by Dermacentor nymphs at 38.95 (95%CI: 10.34-101.48) and Haemaphysalis adults at 36.14 (95%CI: 9.59-94.43). Among Haemaphysalis adults, pan-phlebovirus was positive in 1/50 pools of H. aborensis with an IR of 20 (95%CI: 1.15-92.65), 1/3 pools of H. atherurus with 333.33 (95%CI: 20.74-848.82), and 1/28 pools of H. hystricis with 35.71 (95%CI: 2.06-160.01) (Table 3).
For the pan-flavivirus, 5/659 pools were positive of which 3/50 pools were H. aborensis with an IR of 60 (95%CI: 16.03-153.29) and 2/402 pools were Haemaphysalis nymphs with an IR of 1.63 (95%CI: 0.29-5.29) (Table 3).
For alphavirus, only 1/659 pools of Haemaphysalis nymphs was positive with an IR of 0.67 (95%CI: 0.04-3.23) for the total number of ticks tested and an IR of 0.81 (95%CI: 0.05-3.92) for the total number of Haemaphysalis nymphs tested (Table 3).
Interestingly, when the positivity of arboviral screening is compared between the two collection periods, all nymph ticks positive for pan-phlebovirus were in the sample collected in February 2017 (Figure 1) whereas the pan-flavivirus and pan-alphavirus were in the sample collected in January 2017.
Figure 1: Number of tick pools positive for pan-phlebovirus in January (n=397 pools/481 ticks) and February 2017 (n=262 pools/1,013 ticks).
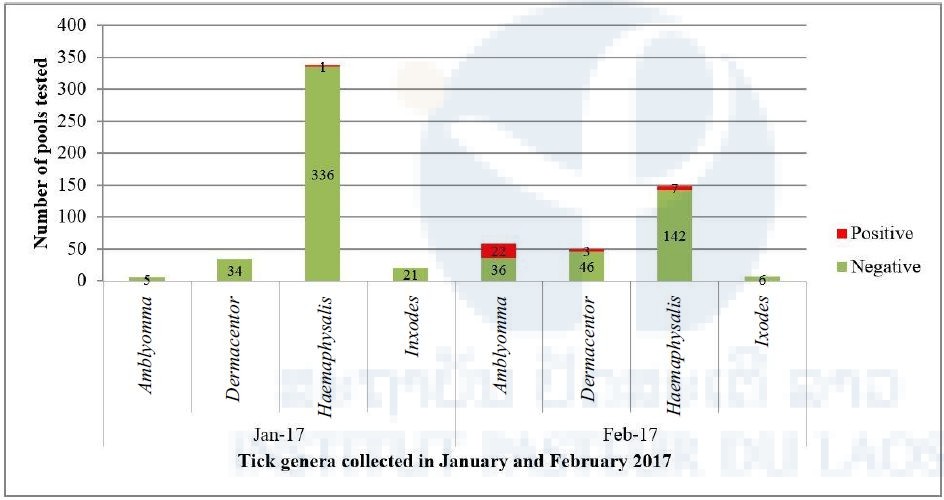
Table 3: Screening of tick specimens for Arboviral sequences
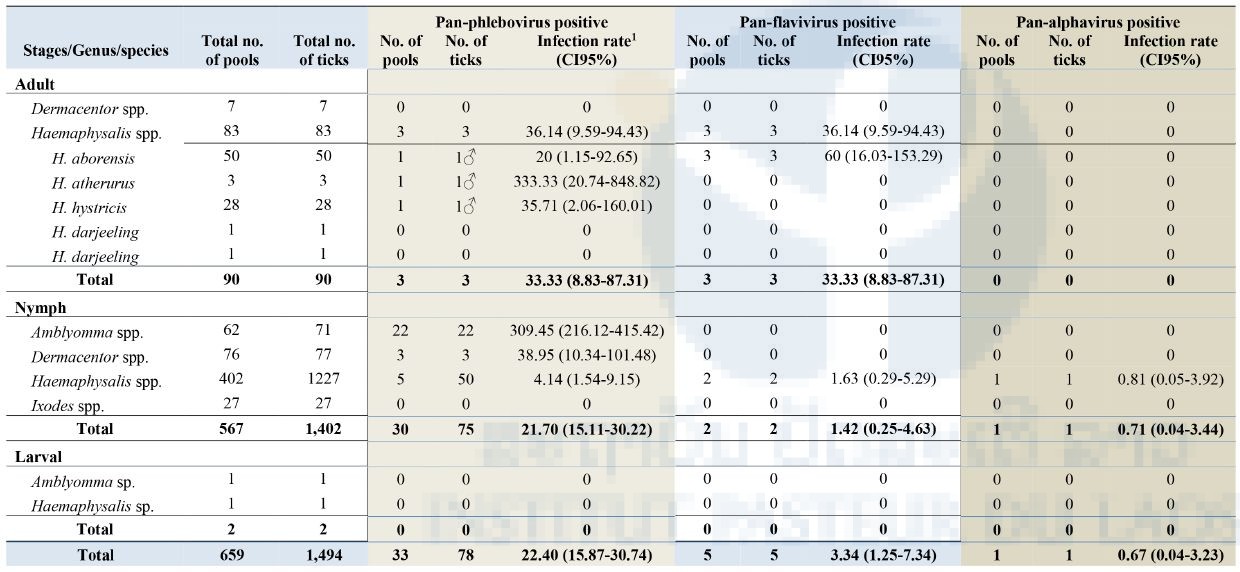
Infection rate (IR) was calculated using Excel add-in software for mosquito surveillance developed by the CDC. Estimates of the IR are presented as the number of infected mosquitoes per 1,000 tested (https://www.cdc.gov/westnile/resourcepages/mosqSurvSoft.html).
Next generation sequencing on tick samples
A first set of 13 positive tick pools was submitted to NGS techniques (Table 4). It was decided to focus on samples found positive by pan-alphavirus RT-PCR due to the potential originality of this finding. In order to reduce technical artifacts, one part of the remaining homogenized tissues was sent in dry ice. Thus, the optimized protocol for the preparation of the nucleic acids could be applied to our samples.
Table 4: NGS analysis of tick samples
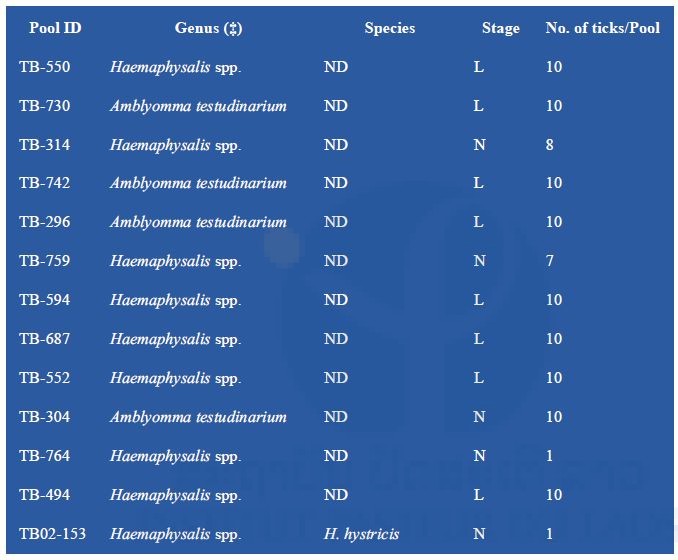
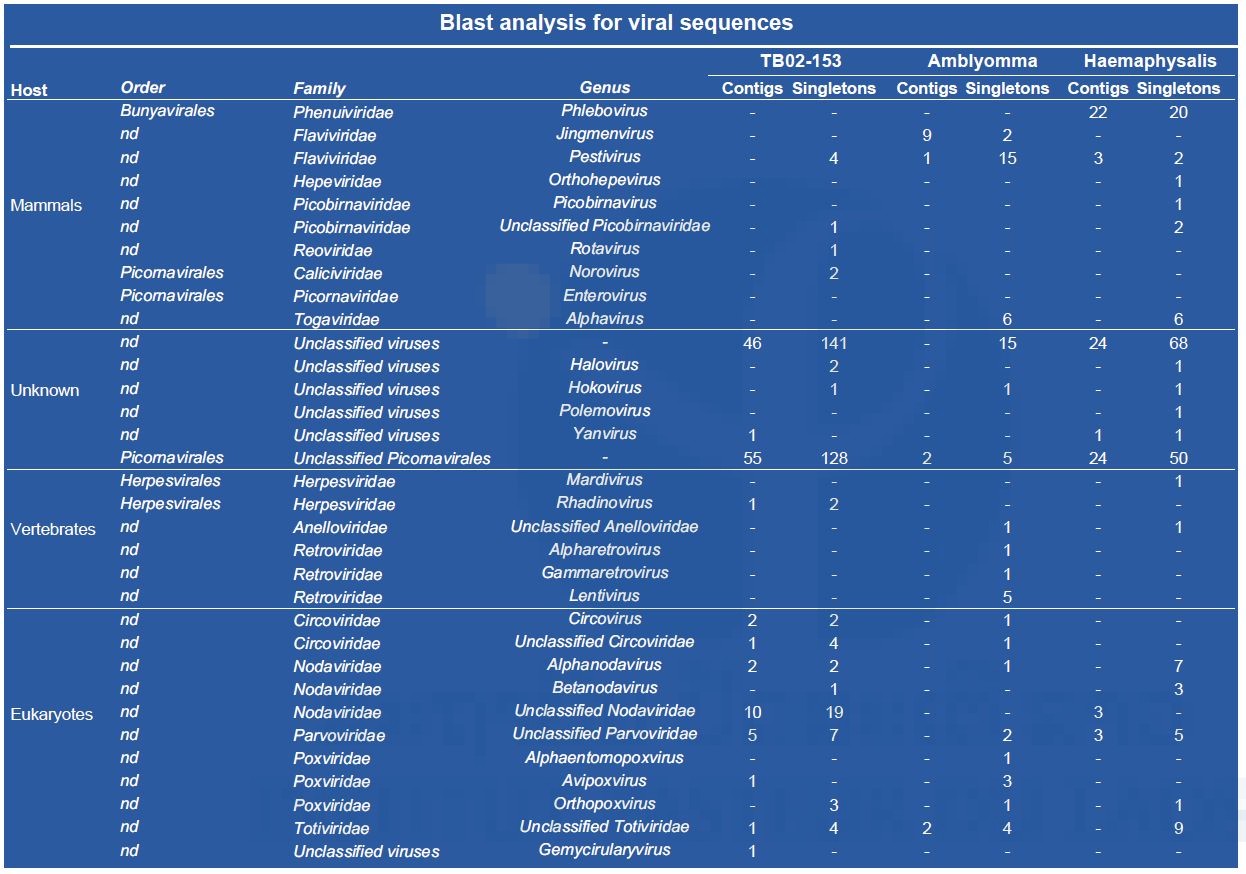
(‡) : specimen gender could not be determined; ND: not determined; L: larvae; N: nymphs To reduce the cost of this first screening, pools of nucleic acid were prepared by tick genus except for TB02-153, which was treated apart. Partial data obtained after a preliminary Blast N analysis of NGS contigs are presented in Table 5. Arbovirus sequences were found in all three nucleic acid preparations.
Table 5: Partial results obtained by Blast analysis of NGS contigs (data related to plants, algae, fungi, etc. are not presented).
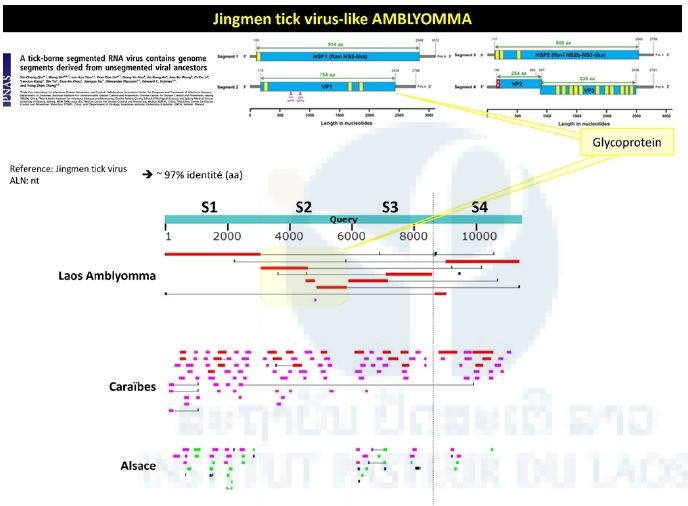
Sequences matching a newly described tick-borne virus (i.e. Jingnen virus) have also been retrieved from the Amblyomma pool. As shown in Figure 2, a strong nucleotide identity level (97%) has been found between the different contigs obtained from the Amblyomma pool and the reference strain of Jingnen virus.
Figure 2: Identification of Jingmen-like sequences from the Amblyomma pool
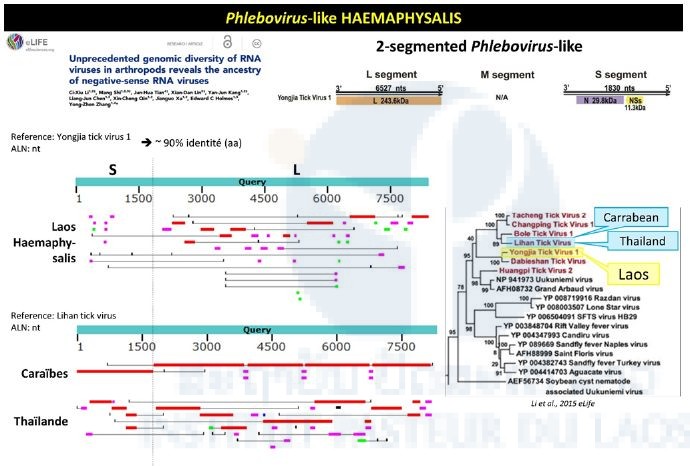
Haemaphysalis pool analysis evidenced partial sequence presenting a 90% nucleotide identity with a group of phlebo-like viruses (Figure 3). Contigs located on the L and S segments matched the reference sequences of the corresponding genome fragments of Yongjia virus, which has also been identified in ticks.
Figure 3: Phlebovirus-like sequences identified in the Haemaphysalis pool
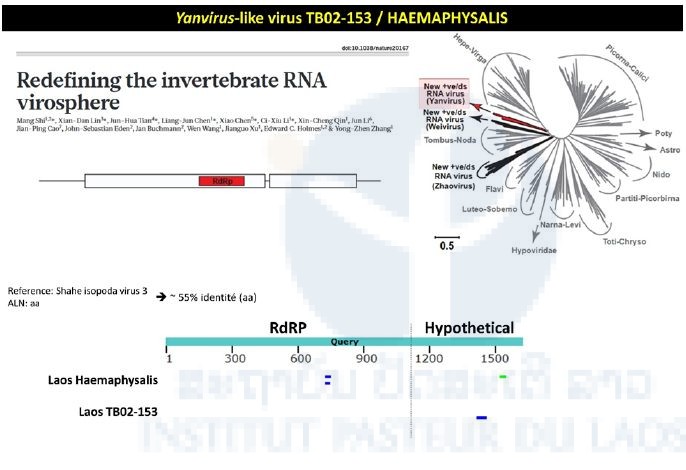
The TB02-153 pool consisted of Haemaphysalis specimens collected in 2013 during the first TickMap project. This pool gave a positive signal when tested by the pan-alpha RT-nested-PCR. Intriguingly, the NGS approach didn’t confirm this preliminary data. Preliminary NGS data revealed matches between the transcriptome of the TB02-153 pool and the RNA dependent RNA polymerase of Yanvirus-like viruses, a new group of viruses recently described (Figure 4).
Figure 4: Yanvirus-like viruses identified in the Haemaphysalis pool
Bacterial screening
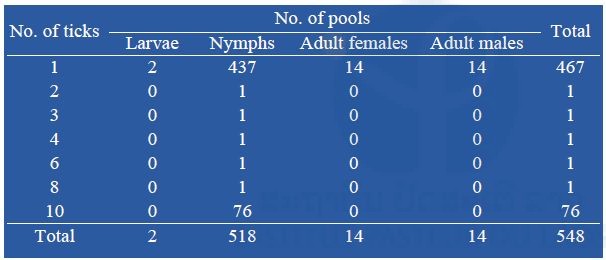
So far, a total of 548 pools containing 1,250 ticks have been screened for the presence of Rickettsia spp., Leptospira spp., Anaplasma spp. (including Anaplasma phagocytophilum), Ehrlichia spp. (including Ehrlichia chaffeensis), Coxiella burnetii and Neorickettsia spp. Almost all the tick pools tested were nymphal stage, totaling 518 pools (437 pools of 1 tick) (Table 6).
Table 6: Number of pooled ticks collected during our surveys submitted to bacterial screening
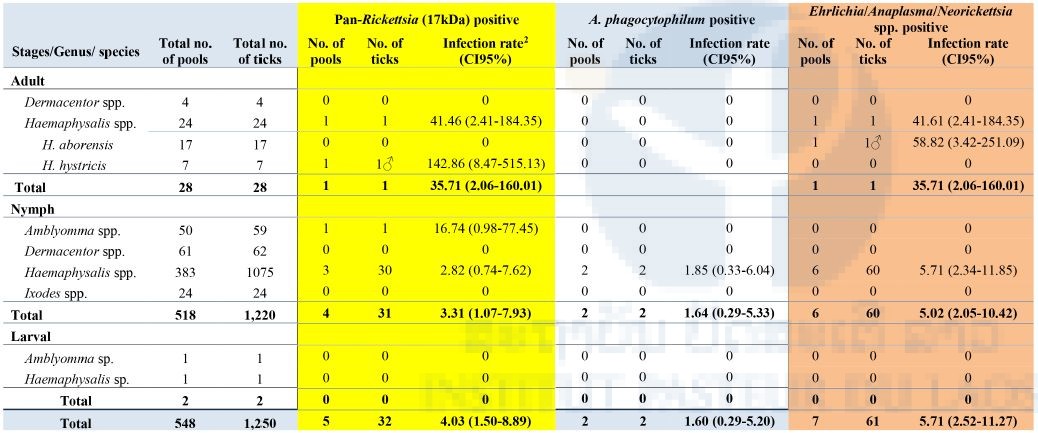
Of the 548 pools analyzed, 5/548 (0.9%) pools were positive for Rickettsia spp. (17kDa) with an infection rate of 4.03 (CI95%: 1.50-8.89) (estimated per 1000 ticks tested), in 1 pool of Haemaphysalis adult (H. hystricis), 3 pools of nymphal Haemaphysalis spp. and 1 pool of nymphal Amblyomma spp. Further analysis and sequencing identified all of these positives as R. raoulti (99% identity). Overall, 7/548 (1.27%) were positive for the Ehrlichia/Anaplasma/Neorickettsia pan-PCR with an infection rate of 5.71 (CI95%: 2.52-11.27) (estimated per 1000 ticks tested) including 1 pool of Haemaphysalis adult: H. aborensis, and 6 pools of nymphal Haemaphysalis spp. Further analysis and sequencing identified six of the positives as Anaplasma bovis and one as an uncultured Anaplasma sp. Overall, only 2/548 pools of nymphal Haemaphysalis spp. were positive for Anaplasma phagocytophilum (Table 7).
None of the tick pools (0%; 0/548) were positive for C. burnetii, Leptospira spp., or Ehrlichia chaffeensis.
In the total of 548 pools (1,250 ticks) that have been tested for all investigated viral and bacterial pathogens, 1 pool of one adult Haemaphysalis hystricis male, 1 pool of ten Haemaphysalis spp. nymphs, and 1 pool of one Amblyomma sp. nymph were positive for pan-phlebovirus and pan-Rickettsia (later identified as R. raoulti).
Table 7: Screening of ticks
2Infection rate (IR) was calculated using Excel add-in software for mosquito surveillance developed by CDC. Estimates of the IR are presented as the number of infected mosquitoes per 1,000 tested (https://www.cdc.gov/westnile/resourcepages/mosqSurvSoft.html).
Molecular tick identification
A total of six samples,1 Dermacentor nymph, 4 Haemaphysalis nymphs, and 1 Ixodes nymph, were tested to obtain a fragment of cytochrome oxidase subunit I (COI), barcode region. Only one sample (Haemaphysalis nymph) got a weak band using the primers LCO/HCO (Fig. 5), we did not get any band using the primers FF1/R1. Hence, the gradient of the annealing temperature was done using the primers LCO/HCO. For the thermocycling protocol of Wang et al., the annealing temperature in the 30 cycles, we tried 45oC – 55oC, the protocol of Rothen et al., 42oC – 52oC, and Foster et al. 50.7oC – 58oC. The agarose gel showed the best annealing temperature at 53.8oC. Unfortunately, when we tried to do a PCR using the annealing temperature at 53.8oC, we did not get any band. For this reason, it was not possible to obtain the fragment of COI barcode.
Figure 5: Gel agarose (1.5%) of a PCR using LCO/HCO primers following Wang et al., Foster et al., and Rothen et al. thermocycling protocols

Discussion
Our project continues to move forward with no currently observed problems or major obstacles. We have successfully completed our two field collections. A total of 1,494 ticks were collected during two field missions. Four genera were identified as Amblyomma spp., Dermacentor spp., Haemaphysalis spp., and Ixodes spp. Adults were identified as Dermacentor auratus (4), D. compactus (1), and D. steini (2); Haemaphysalis aborensis (50); H. atherurus (3), H. darjeeling (1), H. hystricis (28), and H. (Kaiseraina subgenus) sp. (1). Compared to our previous studies in 2014–2015 in the same area (Vongphayloth et al. 2016), at least two new recordings by morphological identification were made in this study: D. compactus and H. atherurus. Similar to our previous surveys in the same area, the nymphal stage was the most abundant, representing 93%, of which 82% was genus Haemaphysalis. By association of nymphs and adults from this and the previous survey in the area, Amblyomma nymph could be identified as A. testudinarium. Unfortunately, for other genera, especially for Dermacentor and Haemaphysalis, the association could not made as many adult species were found and the nymphal stage of some species is unknown. Our attempt to amplify a fragment of cytochrome oxidase subunit I (COI), barcode region in order to sequence it to help in molecular tick identification failed. However, more experiments will be done to try to get the fragment of COI, e.g. tests to get the best thermocycle temperature program and/or design new barcode primers specifically for Laotian ticks. In addition, another gene, 16S ribosomal DNA (rDNA), will also be tested to help in the identification of Laotian ticks. Our morphological and molecular studies for Laotian tick identification will be continuing in collaboration with Richard G. Robbins and Dmitry A. Apanaskevich, United States National Tick Collection, USA.
For arboviral screening, pan-phlebovirus sequences were the most positive (5%) followed by pan-flavivirus (0.7%) and pan-alphavirus (0.1%). The percentage of pan-flavivirus positivity was higher in this study (0.7%) than in our previous studies (0.3%) whereas pan-alphavirus positivity was lower (0.1%) than in previous surveys (2%) (cf. TickMap1 final report 2014). This is the first time that pan-phlebovirus sequences were observed in questing Haemaphysalis and Amblyomma ticks from Laos. Although the pathogenicity of these viruses still unknown, an in-depth study of these viral species’ diversity warrants further investigation as many novel phleboviruses with zoonotic potential isolated from ticks have recently been reported elsewhere e.g. severe fever with thrombocytopenia syndrome virus (SFTSV) and Heartland virus (HRTV) (Yu et al. 2011, McMullan et al. 2012, Wang et al. 2014). The high pan-phlebovirus infection rate was observed in the samples collected in February, especially in Amblyomma ticks. These may result from (i) a low number of Amblyomma ticks having been collected in January, (ii) the viruses having a highly specific host for this tick genus e.g. Heartland virus was first observed from Amblyomma americanum (McMullan et al. 2012), and (iii) the existence of a dynamic vertebrate host population as a pathogen reservoir.
Discrepancies between RT-PCR and NGS results could be explained by the fact that the suspected positivity for alpha sequences only relied on agarose gel electrophoresis analysis. Possible non-specific amplicons may have been detected. Until now, direct sequencing of these PCR products failed as a consequence of the use of degenerated primers designed for large-spectrum amplification within arbovirus genera. New sets of pan-genus primers containing T3 and T7 phage promotor sequences are now under testing to attempt the use of conventional Sanger sequencing to confirm the specificity of the pan-genus amplicons. The preliminary NGS approach revealed a large diversity of viral sequences within the different pools of ticks. The presence of alphavirus sequences in ticks has been confirmed but needs to be consolidated. Original data have also been obtained with new groups of viruses recently found by other groups in tick specimens from Southeast Asia.
Rickettsia spp. was identified in 0.9% of all tick pools. This percentage was low compared to our previous observation (5%) (Taylor et al. 2016). Consistent with the previous finding, Haemaphysalis and Amblyomma were the most positive for Rickettsia spp. Interestingly, 1.27% of 548 tick pools were positive for Neorickettsia spp. This is the first data available on Neorickettsia spp. in Laos and further investigations into their role as causes of animals and human disease may be needed.
Our findings showed that Haemaphysalis spp. (both adult and nymph) and Amblyomma spp. were co-infected by phlebovirus spp. and Rickettsia spp. This is the first data ever available on coinfection by bacteria and virus in Laos. This finding indicated that these pathogens may be vertically transmitted or transmitted by consecutive feedings, coinfected hosts, or cofeeding. It is also possible that the diversity of pathogens (bacteria and viruses) in the area is high. Studies of vertebrate hosts in the area are warranted to provide more information on a possible host reservoir for those pathogens.
References
Folmer, O., M. Black, W. Hoeh, R. Lutz, and R. Vrijenhoek. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294-299.
Foster P. G., E. S. Bergo, B. P. Bourke, T. P. Oliveira, S. S. Nagaki, D. C. Sant’Ana, M. A. M. Sallum. 2013. Phylogenetic analysis and DNA-based species confirmation in Anopheles (Nyssorhynchus) 54063PLoS ONE 8
McMullan, L. K., S. M. Folk, A. J. Kelly, A. MacNeil, C. S. Goldsmith, M. G. Metcalfe, B. C. Batten, C. G. Albarino, S. R. Zaki, P. E. Rollin, W. L. Nicholson, and S. T. Nichol. 2012. A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med 367: 834-841.
Nuttall, G. H. F., W. F. Cooper, C. Warburton, L. E. Robinson, and D. R. Arthur. 1926. Ticks: pt. IV. The genus Amblyomma. Cambridge University Press.
Rothen, J., N. Githaka, E. G. Kanduma, C. Olds, V. Pfluger, S. Mwaura, R. P. Bishop, and C. Daubenberger. 2016. Matrix-assisted laser desorption/ionization time of flight mass spectrometry for comprehensive indexing of East African ixodid tick species. Parasit Vectors 9: 151.
Sanchez-Seco, M. P., D. Rosario, C. Domingo, L. Hernandez, K. Valdes, M. G. Guzman, and A. Tenorio. 2005. Generic RT-nested-PCR for detection of flaviviruses using degenerated primers and internal control followed by sequencing for specific identification. J Virol Methods 126 (1-2): 101-109.
Sanchez-Seco, M. P., D. Rosario, E. Quiroz, G. Guzman and A. Tenorio. 2001. A generic nested-RT-PCR followed by sequencing for detection and identification of members of the alphavirus genus. J Virol Methods 95(1-2): 153-161.
Tanskull, P. and I. Inlao. 1989. Keys to the adult ticks of Haemaphysalis Koch, 1844, in Thailand with notes on changes in taxonomy (Acari: Ixodoidea: Ixodidae). J Med Entomol 26(6): 573–600.
Taylor, A. J., K. Vongphayloth, M. Vongsouvath, M. Grandadam, P. T. Brey, P. N. Newton, I. W. Sutherland and S. Dittrich. 2016. Large-Scale Survey for Tickborne Bacteria, Khammouan Province, Laos. Emerg Infect Dis 22(9): 1635–1639.
Vongphayloth, K., P. T. Brey, R. G. Robbins and I. W. Sutherland. 2016. First survey of the hard tick (Acari: Ixodidae) fauna of Nakai District, Khammouane Province, Laos, and an updated checklist of the ticks of Laos. Syst Appl Acarol 21(2): 166–180.
Wang G., C. Li, X. Guo, D, Xing, Y. Dong, Z. Wang, Y. Zhang, M. Liu, Z. Zheng, H. Zhang, X. Zhu, Z. Wu, T. Zhao. 2012. Identifying the main mosquito species in China based on DNA barcoding. 47051PLoS ONE 7
Wang, J., P. Selleck, M. Yu, W. Ha, C. Rootes, R. Gales, T. Wise, S. Crameri, H. Chen, I. Broz, A. Hyatt, R. Woods, B. Meehan, S. McCullough, and L. F. Wang. 2014. Novel phlebovirus with zoonotic potential isolated from ticks, Australia. Emerg Infect Dis 20: 1040-1043.
Yamaguti N., V. J. Tipton, H. L. Keegan, and S. Toshioka (1971). Ticks of Japan, Korea, and the Ryukyu islands. Brigham Young University Science Bulletin, Biological Series 15(1):1.
Yu, X. J., M. F. Liang, S. Y. Zhang, Y. Liu, J. D. Li, Y. L. Sun, L. Zhang, Q. F. Zhang, V. L. Popov, C. Li, J. Qu, Q. Li, Y. P. Zhang, R. Hai, W. Wu, Q. Wang, F. X. Zhan, X. J. Wang, B. Kan, S. W. Wang, K. L. Wan, H. Q. Jing, J. X. Lu, W. W. Yin, H. Zhou, X. H. Guan, J. F. Liu, Z. Q. Bi, G. H. Liu, J. Ren, H. Wang, Z. Zhao, J. D. Song, J. R. He, T. Wan, J. S. Zhang, X. P. Fu, L. N. Sun, X. P. Dong, Z. J. Feng, W. Z. Yang, T. Hong, Y. Zhang, D. H. Walker, Y. Wang, and D. X. Li. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364: 1523-1532.
Publications
Taylor, A. J., K. Vongphayloth, M. Vongsouvath, M. Grandadam, P. T. Brey, P. N. Newton, I. W. Sutherland, and S. Dittrich. 2016. Large-Scale Survey for Tickborne Bacteria, Khammouan Province, Laos. Emerg. Infect. Dis. 22: 1635-1639.







