Molecular epidemiology of rabies in Lao PDR
 Collaboration
Collaboration
National Animal Health Laboratory (NAHL), Ministry of Agriculture and Forestry, Lao PDR.
Funding
• French Ministry of Higher Education, Research and Innovation (MESRI), France.
Objectives
The objective of this project was to conduct a molecular epidemiology study on approximately 100 rabies strains collected by NAHL between 2017 and 2023 and elucidate the rabies circulation in animals of Lao PDR.
Background
Rabies is a fatal viral zoonotic disease that poses a significant public health threat, particularly in developing countries. Annually, approximately 59,000 people die from rabies, with around 80% of human cases occurring in rural areas (1).
Notably, over 40% of deaths involve children under the age of 15. In Asia, the estimated annual death toll is approximately 38,000, though the actual number may be higher due to poor diagnosis and underreporting in many regions. Dogs are the primary reservoir of rabies, responsible for over 95% of human cases.
In Laos, between 2012 and 2022, the Ministry of Health (MOH) reported an annual incidence of human rabies cases ranging from 2 to 24 (mean = 8) (2), a figure lower than the most recent estimate of 38 deaths per year (1). In order to monitor rabies in animals, the National Animal Health Laboratory (NAHL) initiated rabies surveillance in 2004, utilizing the Direct Fluorescence Antibody Test (DFAT) on brain tissue samples.
Since 2012, no molecular data on rabies strains circulating in animals in Lao PDR has been available (3). To fill this gap, IPL collaborated with NAHL to sequence rabiespositive samples from animals. As part of this initiative, a retrospective analysis of rabies strains from 2017 to 2023 was conducted to better understand the patterns of rabies circulation among animals in Laos.
Methodology
Samples.
A total of 119 brain samples that tested positive for rabies using the Direct Fluorescence Antibody Test (DFAT) were shared by NAHL. The majority of these samples were collected from dogs (n=115), while two samples were obtained from cats and two from cattle (Figure 1).
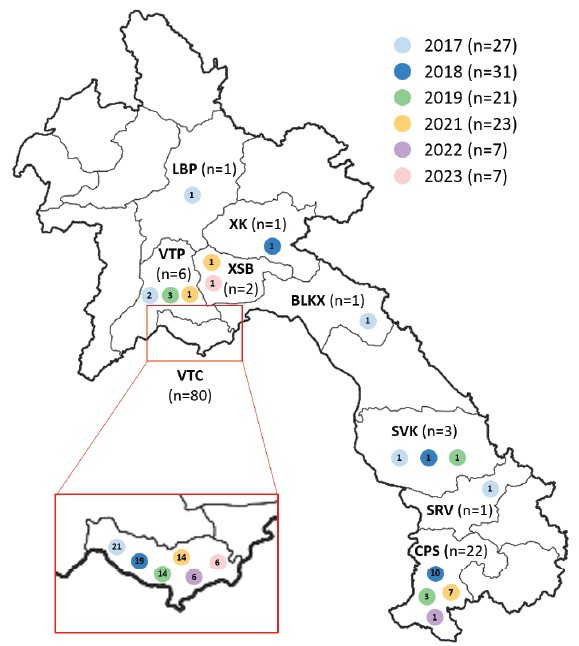
Figure 1: Origin of animal samples positive for rabies obtained from 2017 to 2023.
The number of samples per year is indicated in the coloured circles. The name of provinces is indicated by two-or-three letter code: Borikhamxay (BLKX); Champasack (CPS); Luang Prabang (LBP); Saravane (SRV); Savannakhet (SVK); Vientiane Capital (VTC); Vientiane Province (VTP); Xiengkhuang (XK); Xaysomboune (XSB). The province of origin was not available for two samples.
Rabies virus detection.
Virus nucleic acids are extracted from brain samples by Direct-zol RNA Miniprep kit (Zymo Research) according to the manufacturer’s instructions. Extracted RNAs are then screened for presence of RABV by SYBR Green pan-lyssavirus RT-qPCR assay following WOAH recommendation (4). The RNA extraction quality is validated by SYBR Green RT-qPCR assay targeting an endogenous gene (b-actin).
NGS sequencing.
RNA was then reverse transcribed using LunaScript RT Super Mix (New England Biolabs) according to the manufacturer’s instructions. The complete viral genome (excluding the 3’ and 5’ extremities, corresponding to the leader and the trailer regions, respectively) was amplified with six overlapping PCR fragments by using the Phusion polymerase (ThermoFisher) as previously described (5). Samples were multiplexed using Oxford Nanopore rapid barcode and run in batches of 8-24 on a single R9 flow cell. Basecalling was performed on MinKNOW software. The consensus sequences were then obtained using the EDGE pipeline using the sequence of Lao4 strain (AB981664) as a reference for the mapping step.
Phylogenetic analysis.
All consensus sequences were manually inspected for accuracy, such as the presence of intact open reading frames. A sequence alignment of the newly Lao RABV sequenced genomes combined with the 265 complete genome sequences collected from GenBank was constructed using MAFFT. Maximum likelihood (ML) tree was inferred using IQ Tree web server (http://iqtree. cibiv.univie.ac.at) with automatic substitution model selection using ModelFinder and then edited with FigTree v1.4.4 software (http://tree.bio.ed.ac.uk).
Results
Selection of samples for NGS sequencing.
Out of the 119 samples received at IPL, 11 tested negative for rabies virus (RABV) using the SYBR Green panlyssavirus RT-qPCR assay. Six samples had high cycle threshold (Ct) values (>30), preventing the successful amplification of PCR products, while five samples with low Ct values failed to produce one or more required fragments. Consequently, a total of 97 samples were selected for NGS.
Phylogenetic analysis.
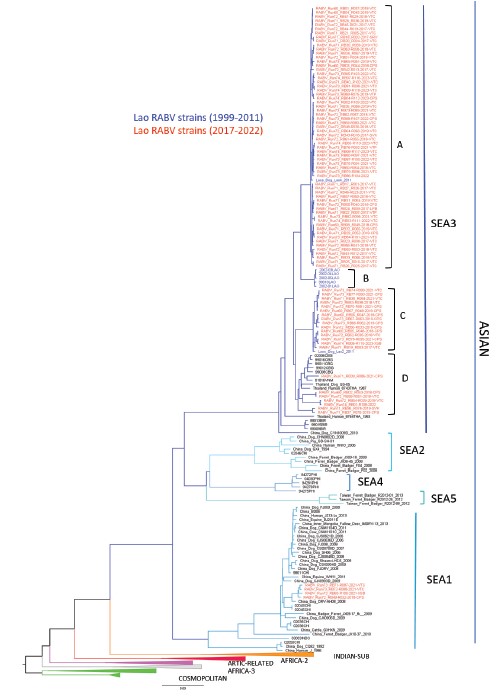
The preliminary phylogenetic analysis revealed that rabies viruses circulating in Laos grouped into several subclades belonging mainly to the Southeast Asia 3 (SEA3) subclade, belonging to the ASIAN clade. Inside the SEA3 subclade, Lao strains grouped in 4 different clusters (A-D) (Figure 2). In addition, some strains showed genetic similarity to viruses from the SEA1 clade, which had mainly been reported in China. More comprehensive findings will be shared in forthcoming publications and reports.
Discussion
Preliminary phylogenetic analysis revealed that rabies virus (RABV) strains circulating in Laos are grouped into several subclades, primarily within the Southeast Asia 3 (SEA3) subclade, which is part of the larger ASIAN clade, as previously described (5). SEA3 subclades also include rabies viruses from Cambodia, Vietnam, and Thailand, suggesting regional cross-border transmission of the virus. Moreover, some strains exhibited genetic similarities to viruses from the SEA1 clade, which had previously been predominantly reported in China (5). This finding indicates potential genetic exchange or common ancestry between rabies viruses in these regions, suggesting a wider geographical distribution of the SEA1 clade than previously documented.
Further analysis will be necessary to gain a deeper understanding of rabies circulation in animals in Laos. This preliminary analysis was based on full-length genome sequences, for which the number of available data in public databases like GenBank is still limited. Additional analyses focusing on the nucleoprotein (N) or glycoprotein (G) genes of RABV, for which more sequences are available in GenBank, should provide better insights into rabies circulation among animals and help study the cross-border transmission of the virus.
Conclusion & perspectives
These preliminary results suggest that certain rabies virus strains may have been introduced into Laos from neighboring countries. This research emphasizes the transboundary nature of rabies circulation in Southeast Asia and highlights the need for ongoing surveillance and cross-border collaboration to effectively manage rabies outbreaks in the region.
References
1. Hampson K, Coudeville L, Lembo T, Sambo M, Kieffer A, Attlan M, et al. Estimating the global burden of endemic canine rabies. PLoS Negl Trop Dis. 2015 Apr;9(4):e0003709. 2. WOAH, ASEAN. ASEAN Rabies Elimination Strategy. World Organisation for Animal Health available at https://asean.org/wp-content/uploads/2017/02/ASEANRabies- Elimination-Strategy.pdf. 2015. 3. Ahmed K, Phommachanh P, Vorachith P, Matsumoto T, Lamaningao P, Mori D, et al. Molecular epidemiology of rabies viruses circulating in two rabies endemic provinces of Laos, 2011-2012: regional diversity in Southeast Asia. PLoS Negl Trop Dis. 2015 Mar;9(3):e0003645. 4. WOAH. CHAPTER 3 .1.18 . RABIES (INFECTION WITH RABIES VIRUS AND OTHER LYSSAVIRUSES). World Organisation for Animal Health, available at: https://www.woah.org/fileadmin/Home/fr/Health_ standards/tahm/3.01.18_RABIES.pdf. 2023. 5. Troupin C, Dacheux L, Tanguy M, Sabeta C, Blanc H, Bouchier C, et al. Large-Scale Phylogenomic Analysis Reveals the Complex Evolutionary History of Rabies Virus in Multiple Carnivore Hosts. PLoS Pathog. 2016 Dec;12(12):e1006041.







