SARS-CoV-2 variants surveillance in Lao PDR
 Collaborations
Collaborations
• National Centre for Laboratory and Epidemiology (NCLE), Ministry of Health, Lao PDR.
• Department of Communicable Diseases Control (DCDC), Ministry of Health, Lao PDR.
Funding
• World Health Organization, Lao PDR country office.
Objectives
The main objective of the SARS-CoV-2 variants surveillance performed at IPL is to generate sequence data on the SARS-CoV-2 strains shared by the NCLE, in order to provide the Lao MOH and the international community (via GISAID) data on the circulation of SARS-CoV-2 variants within Lao PDR as part of the epidemic control strategy.
Background
After the emergence of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) virus in Wuhan, China during late 2019, the Institut Pasteur du Laos (IPL) was requested by the Lao Ministry of Health (MOH) to support the national pandemic response. As a result, since March 2020, IPL has been one of the frontline laboratories for the diagnosis of SARS-CoV-2 in the country. In April 2021, IPL initiated surveillance of circulating SARS-CoV-2 variants through Sanger sequencing and RT-PCR screening. By November 2022, IPL further advanced its efforts by implementing a next-generation sequencing (NGS) method to obtain full-length genome sequences of SARS-CoV-2 variants, enhancing the laboratory’s capacity to monitor viral evolution and variant emergence in the country.
Methodology
Samples.
Respiratory samples were selected by NCLE among the strains identified by the national surveillance and sent to IPL every two weeks.
RT-PCR testing of SARS-CoV-2.
Viral RNA was extracted from clinical specimens using the Nucleo Spin RNA virus kit (Macherey-Nagel) or the Nucleic Acid Extraction Rapid kit (Bioperfectus), according to the manufacturer’s instructions. Extracted RNAs were then screened for the detection of the SARS-CoV-2 genome by RT-PCR following the Charité-Berlin protocol (1).
Next-Generation Sequencing.
At the end of 2022, IPL established a highly multiplexed PCR amplicons approach using the ARTIC Network multiplex PCR primers sets (https://artic.network/ncov- 2019) with the Oxford Nanopore MinION technology (2,3). Samples were multiplexed using Oxford Nanopore rapid barcode and run in batches of 4-24 on a single flow cell. Basecalling was performed on MinKNOW software. The consensus sequences were then obtained using the EDGE pipeline (4). Lineages and specific mutations were determined from consensus sequences using Nextclade (https://clades.nextstrain.org) and PANGOLIN (https:// pangolin.cog-uk.io) web servers. Complete genomes of the SARS-CoV-2 virus generated by IPL were submitted to GISAID (5).
Results
Surveillance of SARS-CoV-2 variants.
Between January and October 2024, NCLE submitted 161 samples from nine provinces to IPL for NGS sequencing (Figure 1). IPL successfully identified SARS-CoV-2 variants for 129 samples (80%). Thirty-two samples could not be identified due to high cycle threshold (Ct) values (>30) or insufficient sequence coverage (<80%) and thus the variant could not be accurately identified using the Nextclade tool.
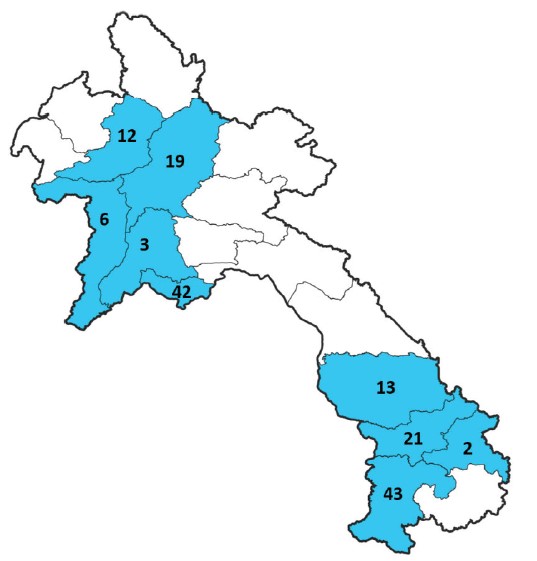
Figure 1: Origin of samples submitted by NCLE to IPL for NGS sequencing.
Most of the identified strains (n=121) were of the Omicron BA.2.86 variant group, classified as a variant of interest (VOI) by the World Health Organization (WHO) on November 21, 2023 (https://www.who.int/ activities/tracking-SARS-CoV-2-variants). This variant was first detected in Lao PDR in December 2023. Six of the identified strains were recombinant variants (XDD, XDV, and XDQ), and two strains were the Omicron subvariant EG.5.1, which circulated in Lao PDR between June 2023 and February 2024.
Within the Omicron BA.2.86 group, 108 samples were identified as the JN.1 variant (and its sub-variants), which WHO classified as a VOI on December 18, 2023, and which has been circulating in Lao PDR since January 2024. Additionally, six samples were identified as the JN.2 variant, which circulated in Laos between December 2023 and March 2024. Several variants under monitoring (VUM), derived from the JN.1 variant, were also detected in Lao PDR in 2024, including: i) KP.2 (n=12), circulating since May 2024; ii) KP.3 (n=22), circulating
since June 2024; and iii) LB.1 (n=2), first detected in Lao PDR in July 2024.
Discussion
The ongoing surveillance of SARS-CoV-2 variants at IPL has provided critical insights into the dynamics of viral transmission and evolution within the country. In 2024, the identification of the Omicron BA.2.86 variant group as predominant among the SARS-CoV-2 strains reflects the changing landscape of SARS-CoV-2 in Lao PDR at the end of 2023, emphasizing the need for robust genomic monitoring. The emergence of variants of interest (VOI), such as JN.1, alongside new variants under monitoring (VUM) like KP.2, KP.3, and LB.1, highlights the adaptive nature of the virus, potentially influenced by population immunity and public health interventions.
These findings underscore the importance of maintaining a comprehensive surveillance network, which is essential for informing public health strategies and controlling future outbreaks. Continued investment in sequencing capabilities and data sharing will be pivotal in understanding and mitigating the impact of SARSCoV- 2 variants on public health in Lao PDR.
Conclusion & perspectives
In 2024, IPL shared the SARS-CoV-2 variants surveillance reports with NCLE, DCDC, and WHO by email and via a virtual database shared between IPL, NCLE, and WHO. Subsequently, IPL also submitted the sequences obtained by NGS on the GISAID database (https://www.gisaid. org). These results allowed the accurate identification of the circulating variants present in Lao PDR, which is essential for the national pandemic control strategy established by MOH.
References
1. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 Jan;25(3):2000045.
2. Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature protocols. 2017 May 24;12(6):1261–76.
3. Itokawa K, Sekizuka T, Hashino M, Tanaka R, Kuroda M. A proposal of alternative primers for the ARTIC Network’s multiplex PCR to improve coverage of SARSCoV- 2 genome sequencing [Internet]. bioRxiv; 2020 [cited 2023 Nov 1]. p. 2020.03.10.985150. Available from: https://www.biorxiv.org/content/10.1101/2020.03.10.98 5150v3
4. Li PE, Lo CC, Anderson JJ, Davenport KW, Bishop- Lilly KA, Xu Y, et al. Enabling the democratization of the genomics revolution with a fully integrated webbased bioinformatics platform. Nucleic Acids Res. 2017 Jan 9;45(1):67–80.
5. Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall. 2017 Jan;1(1):33–46.







